Figures & data
Table 1. The sequence of PLCG2 siRNAs.
Table 2. The primer sequences used in the RT-PCR.
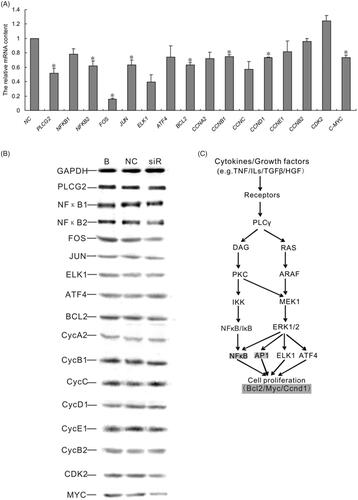
Figure 1. The impact of PLCG2 siRNAs on the mRNA expression level of PLCG2. NC represented cells treated with negative siRNA; siR1, siR2 and siR3 represented cells treated with PLCG2 siRNA 1, siRNA 2 and siRNA 3, respectively. The expression level of PLCG2 mRNA was detected by qRT-PCR.
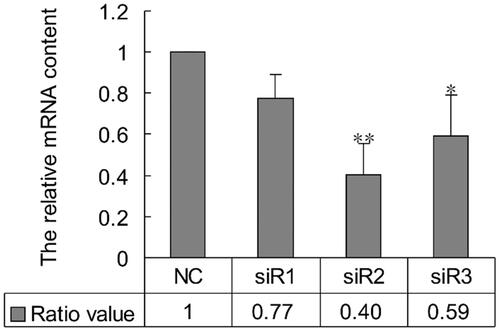
Figure 2. Effect of PLCG2 on cell proliferation. BRL-3A cells were treated by 50 nM siRNA2. At 48 h post-transfection, cells were used in MTT, BrdU and FCM assays to evaluate cell proliferation rate. (A) The results of MTT assay were analyzed using paired Student’s t-test: ** indicates p < .01. The results showed that the viability of cells transfected with siR2 was significantly lower than control (p < .01). (B) Micrograph of the BrdU-labelling cells. Number of BrdU-libeling cells in cells transfected with PLCG2 siRNA2 at 48 h was significantly lower than that of the NC group (39.31 ± 1.12% vs. 45.42 ± 1.06%, *p < .05). (C and E) Cell cycle analysis by flow cytometry. Compared with NC, proportion of cells in S- and G2/M-phase was diminished in cells transfected with siRNA2 at 48 h (S + G2/M: 30.66 ± 2.21% vs.31.92 ± 1.03%, *p < .05). Data represent the mean (±SD) of three independent experiments, at least. (D) BrdU and PI double stained cells to show the proliferative cells. Original magnification: 100×. Scale bar: 100 μm.
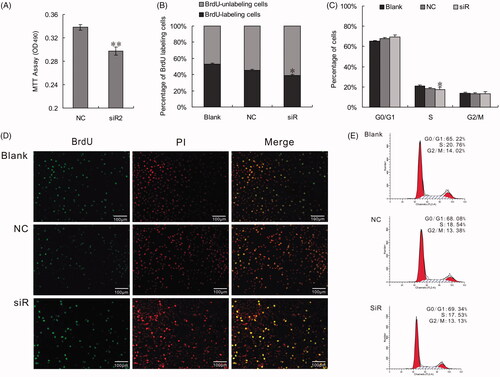
Figure 3. Changes of PLCG2 may modulate NF-κB and ERK signaling, which were hepatocytes proliferation-associated. (A). mRNA expression of 16 genes detected by qRT-PCR. Data represent the mean (±SD) of at least three independent experiments. t-Test: *p < .05. (B) Western blotting assay result shown that the expression of NFKB2, FOS, JUN, BCL2, CCND1 and MYC were suppressed when inhibit the expression of PLCG2. (C) PLCG2 may promote hepatocyte proliferation in vitro via NF-κB and ERK pathway by targeting BCL2, MYC and CCND1.