Figures & data
Figure 1. Isolation of adipose tissue-derived mesenchymal stem cells (AMSCs) and exosomes. (A) The AMSCs surface markers detected by western blots. (B) Representative image of AMSCs. (C) Oil Red O staining in cardiomyocyte administered the adipogenesis treatment. Scale bar = 50 μm. (D) The exosome surface markers detected by western blots. (E) miRNA-320d expression in AMSCs. (F) miRNA-320d expression in exosomes. Values are means ± SEM. For each experiment, sample size n ≥ 3. (*) denotes difference from control group (p < .05).
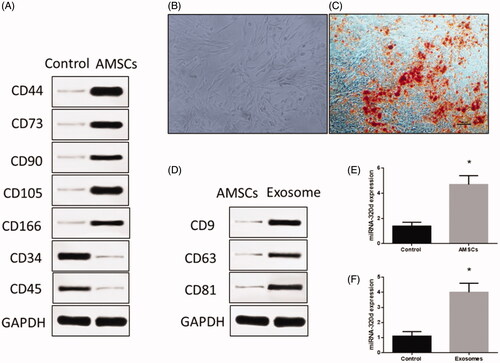
Figure 2. Down-regulation of miR-320d was associated with the regulation of apoptosis and cell viability in cardiomyocytes with AF. (A) Apoptosis level measured by TUNEL staining assays. (B) Apoptosis level measured by Annexin V-FITC/propidium iodide (PI) assays. (C) Cell viability measured by MTT assays. (D) miRNA-320d expression in cardiomyocytes with AF. Values are means ± SEM. For each experiment, sample size n ≥ 3. (*) denotes difference from control group (p < .05).
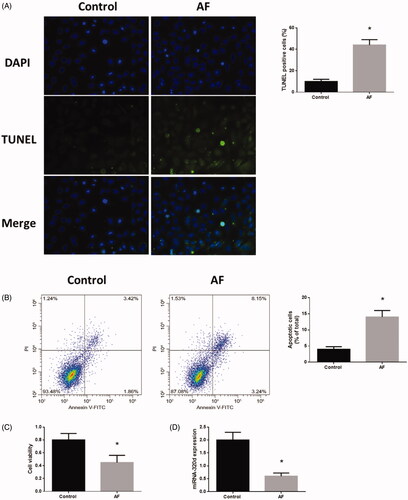
Figure 3. MiR-320d increased in cardiomyocytes co-cultured with modified exosomes. (A) miRNA-320d expression in co-cultured modified exosomes. (B) Apoptosis level measured by TUNEL staining assays. (C) Apoptosis level measured by Annexin V-FITC/propidium iodide (PI) assays. (D) Cell viability measured by MTT assays. (E) Apoptotic protein expressions detected by western blots. Values are means ± SEM. For each experiment, sample size n ≥ 3. (*) denotes difference from control group (p < .05).
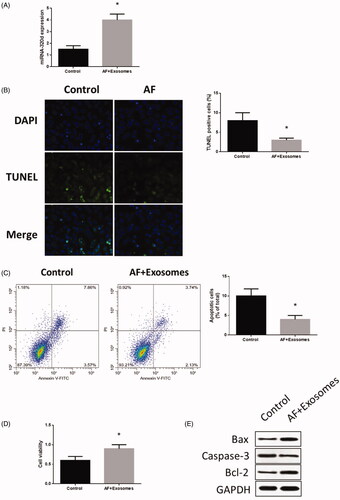
Figure 4. MiR-320d targeted STAT3 in cardiomyocytes with AF. (A) STAT3 mRNA expression detected by real-time PCR. (B) STAT3 protein expression detected by western blots. (C) The predictive binding sequence of miR-320d in the 3′-UTR of STAT3. (D) Relative luciferase activity. Values are means ± SEM. For each experiment, sample size n ≥ 3. (*) denotes difference from control group (p < .05).
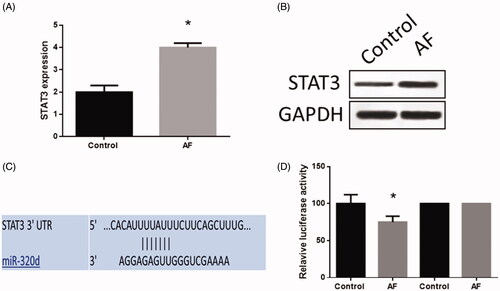
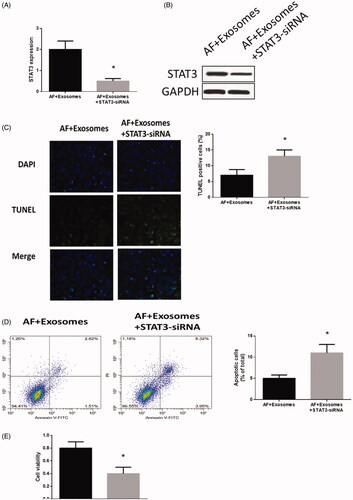
Figure 5. Down-regulation of STAT3 regulated apoptosis and cell viability in cardiomyocytes with AF. (A) STAT3 mRNA expression detected by real-time PCR. (B) STAT3 protein expression detected by western blots. (C) Apoptosis levels measured by TUNEL staining assays. (D) Apoptosis level measured by Annexin V-FITC/propidium iodide (PI) assays. (E) Cell viability measured by MTT assays. Values are means ± SEM. For each experiment, sample size n ≥ 3. (*) denotes difference from control group (p < .05).