Figures & data
Figure 1. Upregulaed circ-PRMT5 was identified in GC tissues and acted as an independent prognostic factor for poor OS in GC patients. (A) circ-PRMT5 expression was statistically significantly higher in GC tissues than in paired normal adjacent tissues from 90 GC patients tested by qRT-PCR; (B) circ-PRMT5 expression was statistically significantly higher in the GC cells (AGS, MKN-28, MKN-45, BGC-823, MGC803, SGC-7901) than in the normal human gastric epithelial GES-1 cells tested by qRT-PCR (p < .05); (C) Kaplan–Meier analysis of the association between the circ-PRMT5 expression levels with the OS of GC patients. The 90 patients were divided into circ-PRMT5 low expression group (n = 45) and circ-PRMT5 high expression group (n = 45) based on the median expression value of circ-PRMT5 in GC tissues, then the Kaplan–Meier survival curves were used to compare the OS between the two groups. *p < .05; **p < .01; ***p < .001.
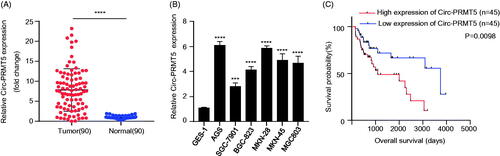
Table 1. Association between circ-PRMT5 expression and GC clinicopathological characteristics in GC patients were analyzed by chi-square test.
Figure 2. Circ-PRMT5 promoted viability, colony formation, migration, invasion and inhibited apoptosis in GC cells. (A) qRT-PCR analysis confirmed that circ-PRMT5 was successfully silenced with si-circPRMT5#1 or si-circPRMT5#2 containing lentivirus infection; the following indicators were then assessed in the AGS and MKN-45 cells infected with the si-NC, si-CircPRMT5#1 or si-CircPRMT5#2 containing lentivirus: (B) cell viability, (C) clone formation assays, (D) cell apoptosis, (E) cell migration, and (F) cell invasion. *p < .05; **p < .01; ***p < .001.
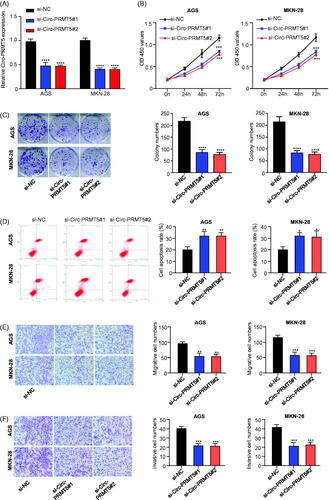
Figure 3. Identification of the potential miRNAs sponged by circ-PRMT5. (A) Sublocalization of circ-PRMT5 was determined by detecting the circ-PRMT5 expression levels in the cytoplasmic and nuclear RNA extraction from AGS and MKN-28 cells, U6 and GAPDH are used as the internal reference in the nucleus and cytoplasm, respectively; (B) Schematic representation of the potential binding sites of miR-145 and miR-1304 with circ-PRMT5 (https://circinteractome.nia.nih.gov/index.html); (C) double luciferase reporter assay in the AGS and MKN-28 cells co-infected with circ-PRMT5 WT or mutated reporter and miR-145 mimics; (D) double luciferase reporter assay in the AGS and MKN-28 cells co-infected with circ-PRMT5 WT or mutated reporter and miR-1304 mimics; (E) Circ-PRMT5 in GC cell lysates was pulled down and enriched with biotin-labelled circ-PRMT5-specific probe; (F) the expression levels of miR-145 and miR-1304 in circ-PRMT5 silenced AGS and MKN-28 cells detected by qRT-PCR; (G) the expression levels of miR-145 and miR-1304 in paired GC and adjacent normal tissues from 90 GC patients (same samples as in ) detected by qRT-PCR; (H) Pearson correlation analysis of the association between miR-145/miR-1304 with circ-PRMT5.
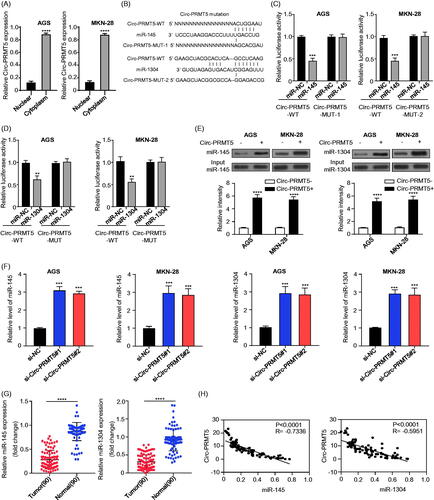
Figure 4. MYC was the downstream target of miR-145 and miR-1304 in GC cells. (A) Schematic representation of the potential binding sites of miR-145 and miR-1304 with MYC-3′UTR (http://www.microrna.org/); (B) double luciferase reporter assay in the AGS and MKN-28 cells infected with WT or mutated MYC-3′UTR reporter and miR-145 or miR-1304 mimics; (C) the expression levels of MYC mRNA in AGS and MKN-28 cells after knockdown (inhibitor)/overexpression (mimic) of miR-145 or miR-1304 detected by qRT-PCR; (D) the protein expression levels of MYC in AGS and MKN-28 cells after knockdown (inhibitor)/overexpression (mimic) of miR-145 or miR-1304 detected by Western-Blot; (E)The expression levels of MYC in paired GC and adjacent normal tissues from 90 patients (same samples as in ) detected by qRT-PCR; (F) Pearson correlation analysis of the association between miR-145/miR-1304 with MYC.
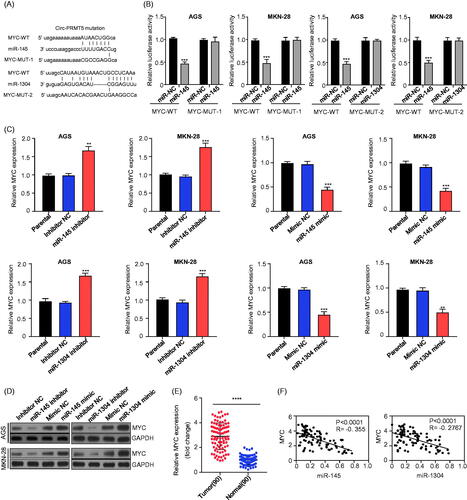
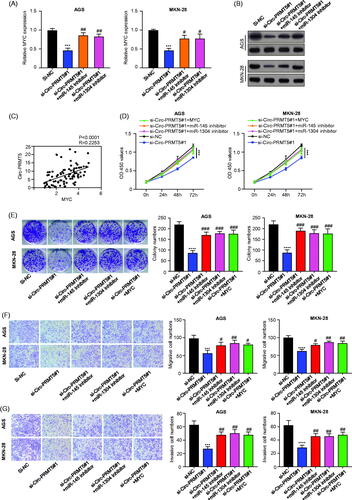
Figure 5. Circ-PRMT5 promoted GC progression by regulation of miR-145/1304/MYC axis. (A) The expression levels of MYC mRNA in AGS and MKN-28 cells with the infection of si-NC, si-circPRMT5#1, si-circPRMT5#1 + miR-145 inhibitor or si-circ-PRMT5#1 + miR-1304 inhibitor detected by qRT-PCR; (B) the expression levels of MYC protein in AGS and MKN-28 cells with the infection of si-NC, si-circ-PRMT5#1, si-circ-PRMT5#1 + miR-145 inhibitor or si-circ-PRMT5#1 + miR-1304 inhibitor detected by Western-Blotting; (C) Pearson correlation analysis of the association between circ-PRMT5 with MYC in GC tissues from 90 patients (same samples as in ); (D) cell viability in AGS and MKN-28 cells with the infection of si-NC, si-circ-PRMT5#1, si-circ-PRMT5#1 + miR-145 inhibitor, si-circ-PRMT5#1 + miR-1304 inhibitor or si-circ-PRMT5#1 + MYC, at different time points, detected by CCK-8 assay; (E) Clone formation assay in AGS and MKN-28 cells with the infection of si-NC, si-circ-PRMT5#1, si-circ-PRMT5#1 + miR-145 inhibitor, si-circ-PRMT5#1 + miR-1304 inhibitor or si-circ-PRMT5#1 + MYC; (F) cell migration ability in AGS and MKN-28 cells with the infection of si-NC, si-circ-PRMT5#1, si-circ-PRMT5#1 + miR-145 inhibitor, si-circ-PRMT5#1 + miR-1304 inhibitor or si-circ-PRMT5#1 + MYC detected by Transwell assay; (G) cell invasion ability in AGS and MKN-28 cells with the infection of si-NC, si-circ-PRMT5#1, si-circ-PRMT5#1 + miR-145 inhibitor, si-circ-PRMT5#1 + miR-1304 inhibitor or si-circ-PRMT5#1 + MYC detected by Transwell with matrigel.