Figures & data
Figure 1. General introduction to MERS-CoV: model structure, life cycle and genomic composition. (A) Cartoon model structure of MERS-CoV. (B) Membrane fusion mechanism for MERS-CoV spike glycoprotein. Binding between RBD and the cell receptor (DPP4) triggers the conformational change of S glycoprotein to form a pre-hairpin intermediate of S2, in which the hydrophobic HR1 is exposed and the fusion peptide inserts into the target cell membrane. This transient S2 intermediate then refolds with HR2 into a stabilized trimer of hairpins, also called six-helix bundle structure (6-HB), bringing the target cell membrane into close proximity of the viral envelope and resulting in the completion of the fusion process. (C) Genomic composition of MERS-CoV. Each coloured box (length in scale) represents one open reading frame in the genomic RNA. The schematic for spike glycoprotein was also shown with labelled domain names and residue numbers. ORF (open reading frame), DPP4 (dipeptidyl peptidase 4), RBD (receptor-binding domain), NTD (N-terminal domain), CTD (C-terminal domain), FP (fusion peptide), and HR1-2 (heptad repeats 1-2).
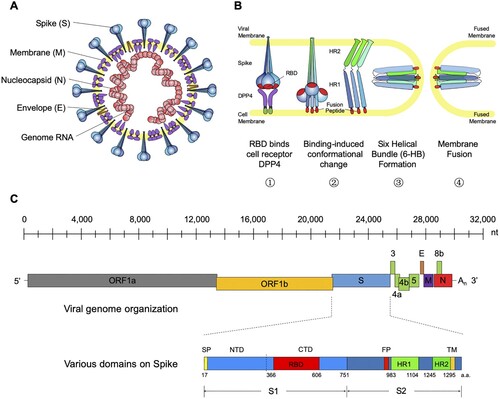
Figure 2. Structural insights of the MERS-CoV spike glycoprotein. (A–B) Top and side view of the MERS-CoV spike trimer with all RBD in ‘down' position, shown as molecular surface (PDB ID: 5W9J). The three protomers are coloured green, lightblue, and red, respectively. The labels are the same with those in . (C) One of the three protomers in (B) is highlighted as cartoon representation whereas the other two protomers are faded in white. The RBD in ‘down' position is coloured in green. The non-RBD S1 region was coloured deep blue and the S2 region was coloured orange. (D) Superimposition of RBD-bound DPP4 (PDB ID: 4L72) into the MERS-CoV spike trimer. Clashes were observed between DPP4 and the other two S1 regions in the trimer structure. (E–F) Top and side view of the MERS-CoV spike trimer with one RBD in ‘up' position and the other two in ‘down' position, shown as molecular surface (PDB ID: 5W9H). Same colour codes are used as in (A–B). (G) The protomer with RBD in ‘up' position is highlighted as cartoon representation, with RBD in green, non-RBD S1 region in deep blue, and S2 in orange. (H) Superimposition of the RBD-bound DPP4 into the MERS-CoV spike trimer, with DPP4 interacting with the ‘up' RBD. No steric clash was observed.
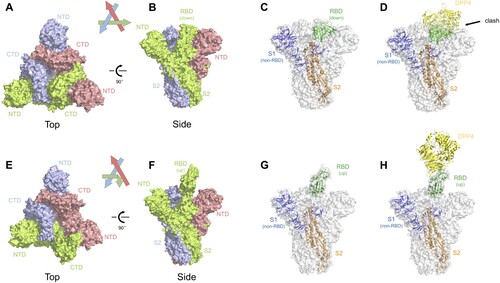
Figure 3. Development of monoclonal antibodies against MERS-CoV. (A) Monoclonal antibodies sorted from non-immunized human scFv (single-chain fragment variable) libraries. MERS-4 and MERS-27 were isolated from a non-immunized human scFv library displayed on yeast with MERS-CoV spike RBD as bait protein. Similarly, 3B11 and m336 were isolated from non-immunized human scFv phage libraries with MERS-CoV S protein or RBD protein as bait protein, respectively. (B) Monoclonal antibodies sorted from immunized animals. The antibodies 5F9, hMS-1 (Mersmab-1), D12, F11, G2, G4, 4C2h (4C2), REGN3048 and REGN3051 were isolated from mice immunized with the indicated vaccines labelled in the colour-coded boxes, each representing a different immunogen; the bait or target protein for antibody selection were also listed. The mice from which REGN3048 and REGN3051 were isolated were given the pale blue colour to indicate that they express human immunoglobulin genes. NbMS10-Fc, JC57-11, JC57-13, JC57-14, F1B-H1 and HCAb-83 were isolated from larger animal including llama, rhesus macaque and camels as indicated. The vaccines and selection criteria were also shown. NTD (N-terminal domain), Fc (fragment constant). (C) Monoclonal antibodies isolated from human survivors recovered from MERS-CoV infection. MERS-GD27, MERS-GD33, LCA60, CDC-C2, CDC-C5, CDC-A2 and CDC-A10 were generated by culturing B cells sorted from the patient and screening for MERS-CoV-specific antibodies. MCA1 was produced by constructing a phage library displaying scFv cloned from a convalescent patient.
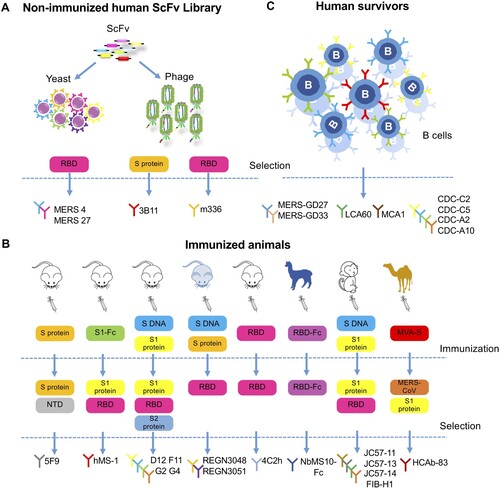
Table 1. Advancement in MERS-CoV monoclonal antibodies development.
Figure 4. Advancement in structural studies of MERS-CoV neutralizing monoclonal antibodies. (A) Structure of MERS-CoV spike trimer ectodomain (PDB ID: 5X5F). A single protomer of the trimeric spike protein with RBD in ‘up' conformation is shown as molecular surface. The RBD, NTD and S2 subunit are coloured in green, paleyellow and lightblue, respectively. The two remaining protomers with RBD in ‘down' conformation are shown in cartoon representation and coloured in wheat. (B) Structures of MERS-CoV neutralizing antibodies targeting RBD. Antibodies are classified into three groups and shown as cartoon representation. The RBD is coloured in green and antibodies in different colours. Group 1 includes MERS-27 (PDB ID: 4ZS6), D12 (PDB ID: 4ZPT), 4C2 (PDB ID: 5DO2) and JC57-14 (PDB ID: 6C6Y). Group 2 includes m336 (PDB ID: 4XAK), MCA1 (PDB ID: 5GMQ), CDC2-C2 (PDB ID: 6C6Z) and MERS-GD27. Group 3 includes MERS-4 (PDB ID: 5ZXV) and MERS-4V2 (PDB ID: 5YY5). (C) Neutralizing mechanisms of MERS-CoV neutralizing antibodies targeting RBD. The left panel shows the structural superimposition of the representative antibodies from the three groups (MERS-27, m336 and MERS-4) and DPP4 (coloured in yellow) bound to RBD (coloured in green, PDB ID: 4L72) at the same time. The right panel is enlarged view of steric clashes between the antibodies and the DPP4 and a significant conformational difference in the RBD β5-β6 loop between antibody-bound and DPP4-bound states. (D) Structure of G4 fab (coloured in teal) in complex with spike trimer (PDB ID: 5W9H). (E) Side views of spike monomer bound to G4 fab. The enlarged view of the glycosylated loop in the S2 subunit recognized by G4 is shown on the right.