Figures & data
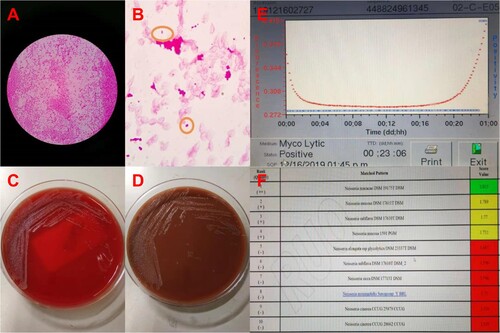
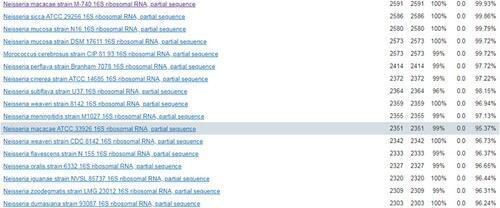
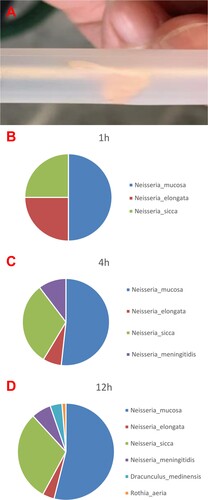
Figure 1. The blood culture was positive for N. macacae in 24 h. (A) Bacterial colony smear (Gram staining). (B) Blood culture was positive, then direct smear (Gram staining). (C) Blood plate for 72 h. (D) Chocolate plate for 72 h. (E) Time curve of blood bottle positive report. The blood culture was positive at 23 h 6 min. (F) The strain from blood culture was identified using MALDITOF MS.