Figures & data
Table 1. Pediatric patients’ characteristics.
Figure 1. Procalcitonin (Panel A) and C-reactive protein (Panel B) kinetics in the four paediatric patients receiving the platelet concentrates. All patients received platelet concentrates on 2 September 2020 except patient 1 who received the concentrates on 1 September 2020. Isolation of L. garvieae occurred for all patients (patients 1, 2, and 3) on 3 September 2020. Cut-off: normal value. Procalcitonin normal value <0.05 ng/mL; C-reactive protein normal value <0.50 mg/dL.
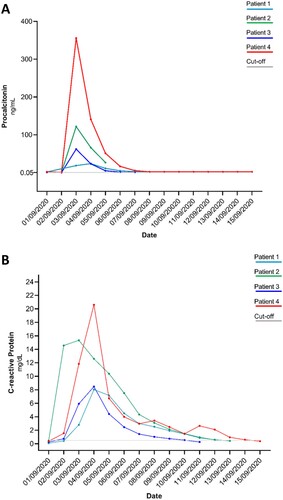
Figure 2. Estimated maximum likelihood (A) and Bayesian (B) phylogenies based on coreSNP genome of the three L. garvieae strains isolated from pediatric patients at Bambino Gesù Pediatric Hospital, IRCCS. Representative 20 L. garvieae and 4 L. lactis strains retrieved by public databases were also included to obtain an alignment of 27 coreSNP genomes 62,990 nucleotides long. The L. lactis and L. garvieae strains were annotated according to collection source (Orange: Dairy products, Blue: Fishes, Brown: Cow, Red: Humans; Other: Purple). BioSample accession numbers defined the strains. Panel (A) reported the ML tree, inferred using iqTree2 under the nucleotide substitution GTR+I+G4 model and 1000 bootstrap replicates. Panel (B) reported the Bayesian tree, performed by BEAST v.1.10.4 setting a chain length of 100 million states under a strict molecular clock model and the GTR+I+G4 substitution model. Bootstrap values and posterior probabilities are reported along the branches of the trees.
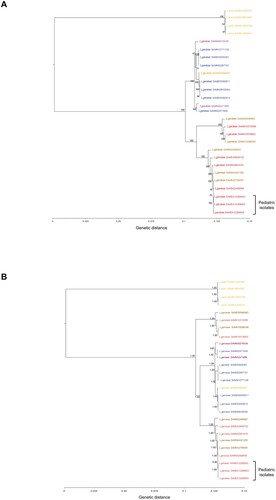
Table 2. Putative virulence factors and resistance genes identified in the three L. garvieae strains isolated from pediatric patients at Bambino Gesù Pediatric Hospital, IRCCS.
