Figures & data
Figure 1. Characteristics and phylogenetic groups distribution, and maximum clade credibility (MCC) trees of PG1 and PG3 clades. A. Serotype distribution of the isolates used in this study. The inner, middle, and outer circles depict the proportion of human isolates from China, animal isolates, and human isolates from other countries by serotype. Serotypes were based on genome sequence predictions. Y-gtrI, Y-gtrIII and Yv-gtrII denotes the gtrI, gtrIII and gtrII genes detected in these isolates. B. Phylogenetic group (PG) distribution of the isolates used in this study. The inner, middle and outer circles depict the proportion of human isolates from China, animal isolates, and human isolates from other countries by the PGs. C: Maximum clade credibility (MCC) tree of PG1 isolates. The branches of animal isolates lineages are coloured in blue. For details of the phylogenetic relationships among the isolates, see supplementary figure S1. D: Maximum clade credibility tree of PG3 isolates. The branches of animal isolate lineages are coloured in blue. For details of the phylogenetic relationships of the isolates, see supplementary Figure S2.
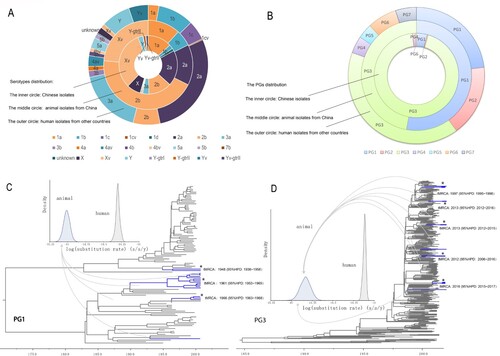
Figure 2. Population structure of the 648 S. flexneri isolates. A: PGs of the 648 S. flexneri isolates. The PGs were identified from the maximum likelihood tree of the 648 isolates and nodes within the PGs are collapsed. B: Exploration of the temporal signal in the data by regressing root-to-tip genetic distances against sampling times with residuals coloured by PG. Correspondence between the colours of the residuals and PGs is as indicated in the legend. The numbers between brackets indicate the strains obtained for this study in China from human and animal hosts, respectively for which information regarding sampling time was available. C: Maximum-likelihood tree of S. flexneri inferred from 61,581 single nucleotide polymorphisms (SNPs). All SNPs were recorded by their position in reference to the 2002017 genome. Potential genome-wide SNPs outside the recombinant regions were used. Coloured rings from the inside to outside indicate the study origin, geographical regions, and PG groups and hosts.
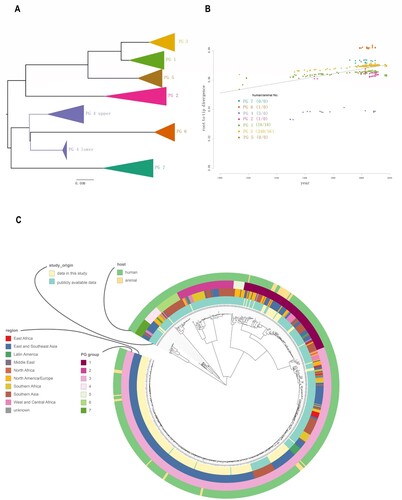
Figure 3. Clade-wise exploration of the temporal signals of PG1 and PG3 Root to tip divergence was computed using TempEST, with and without the animal isolates for PG1 and PG3 considered separately. The animal and human isolates are coloured as shown.
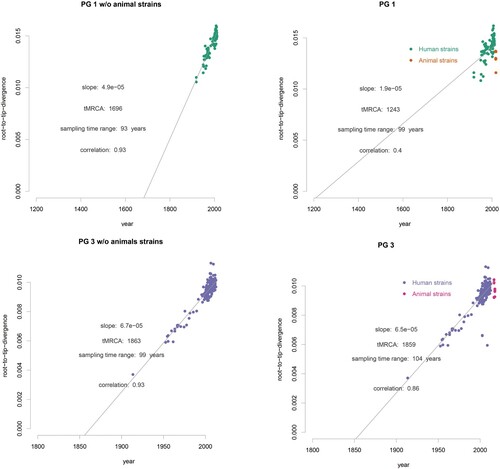
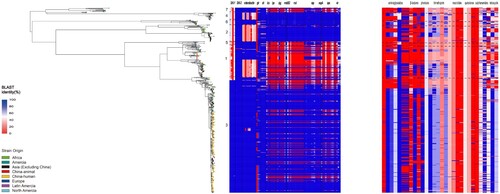
Figure 4. Comparison of the distribution of virulence factors and antimicrobial resistance determinants of S. flexneri isolates from animals and Chinese human isolates within and between PG1 and PG3. The compositions of virulence factors and AMR determinants in each isolate are represented as the percentage identity of the best BLAST hit. The virulence factors/genes (in order) are SHI-1 (pic, set1A, set1B, and sigA), SHI-2 (iucA, iucB, iucC, iucD, and iutA), enterobactin genes (entA, entB, entD, entE,entF, fepA, fepB, fepC, fepD, and fepG), sit (sitA, sitB, sitC, and sitD), ics (icsA/virG, icsB, and icsP), ipa (ipaA, ipaB, ipaC, and ipaD), ipg (ipgA, ipgB1, ipgB2, ipgC, ipgD, ipgE, and ipgF), msbB2, mxi (mxiA, mxiC, mxiD, mxiE, mxiG, mxiH, mxiI, mxiJ, mxiK, mxiL, mxiM, and mxiN), osp (ospB, ospC1, ospC2, ospC3, ospC4, ospD1, ospD2, ospD3, ospE1, ospE2, ospF, and ospG), sepA, spa (spa13, spa15, spa24, spa29, spa32, spa33, spa40, spa47, and spa9), and vir (virA, virB, virF, and virK). The AMR genes are aac(3)-II, aadA1, aadA2, aadA5, strA, strB, and sat1 (aminoglycosides); blaCTX-M-24, blaOXA-1, and blaTEM-1 (β-lactams); catA1 and catB1 (phenicols); dfrA17, dfrA3b, dfrA1, dfrA5, dfrA14 and dfrA8 (trimethoprims); ermB, msrE, mphA,and mphE (macrolides); qacEΔ1 and qnrS1(quinolones); qepA, sul1, and sul2 (sulphonamides), and tetA(A), tetA(D), and tetA(B) (tetracyclines).
Supplemental Material
Download Zip (2.1 MB)Data availability
Sequencing data were submitted to the Sequence Read Archive (SRA) with the project number PRJNA820478 (The strains used are listed in Supplementary file 1).
