Figures & data
Figure 1. ASFV DNA copies in the various organ tissues (approximately 1 g) collected from pigs in the ASFV infection group. Red bars indicate the mean value in each tested organ tissue. *p < 0.05. LN: lymph node; ASFV, African swine fever virus; dpi, days post-infection
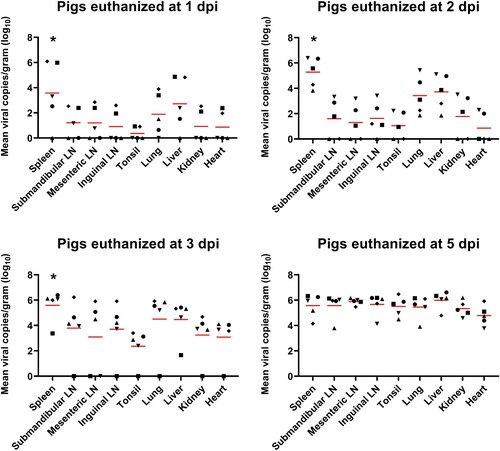
Table 1. List of overlapping DEGs in the nine organ tissues obtained from ASFV-infected pigs euthanized at 1 and 3 dpi.
Figure 2. Heatmap plots representing the expression profile of DEGs involved in the different immune responses in each organ tissue from the ASFV-inoculated pigs (TRT pigs) euthanized at 1 and 3 dpi. SLN, submandibular lymph node; MLN, mesenteric lymph node; ILN, inguinal lymph node; DEGs, differentially expressed genes; ASFV, African swine fever virus
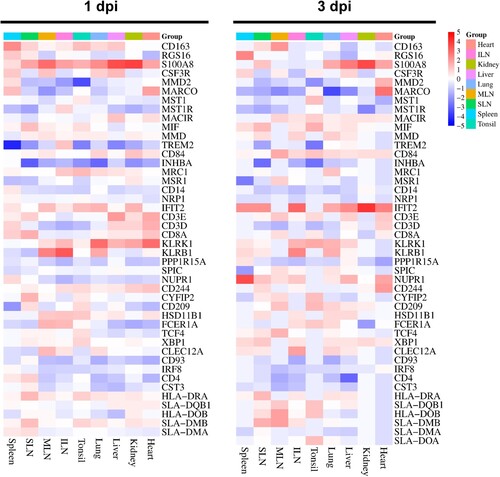
Figure 3. Gene expression patterns of cytokines and their receptors in organ tissues (tonsils, spleen, SLN, MLN, lungs, liver, kidneys, ILN, and heart) from ASFV-inoculated pigs (TRT) euthanized at 3 dpi. (A) ILs and their receptors. (B) IFNs. (C) Genes of the TNF-receptor superfamily. (D) Chemokines and their receptors. SLN, submandibular lymph node; MLN, mesenteric lymph node; ILN, inguinal lymph node; TNF, tumour necrosis factor; ASFV, African swine fever virus; IFN, interferon; IL, interleukin
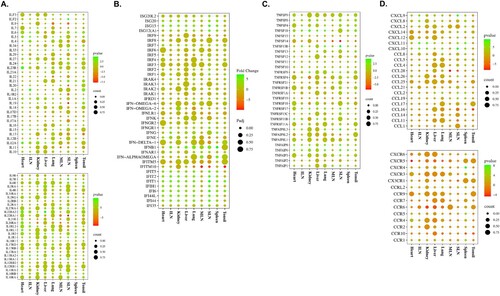
Table 2. List of overlapping immune-associated pathways that were significantly (p < 0.05) enriched with DEGs identified in organ tissues from ASFV-infected pigs, at 1 and 3 dpi
Figure 4. Illustration of the conserved immune-related pathways in organ tissues (the spleen, SLN, MLN, ILN, tonsils, lungs, liver, kidneys, and heart) from ASFV-inoculated pigs (TRT) euthanized at (A) 1 and (B) 3 dpi. The KEGG pathways terms are listed on the left, and p-value and gene counts are shown on the right. SLN, submandibular lymph node; MLN, mesenteric lymph node; ILN, inguinal lymph node; KEGG, Kyoto Encyclopedia and Gene and Genomes; ASFV, African swine fever virus; IL, interleukin; PI3 K, phosphoinositide 3-kinase; NF-κB, nuclear factor-kappa B; dpi, days post-inoculation
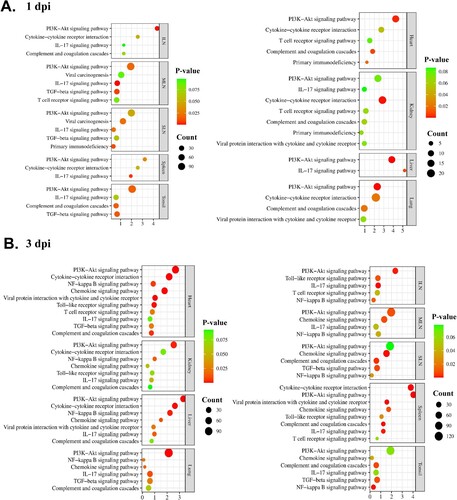
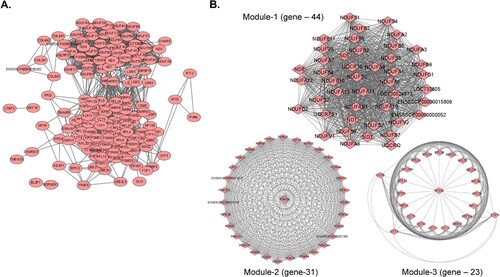
Figure 5. (A) PPI network for the 75 common DEGs identified in the nine organ tissues (spleen, submandibular lymph node, mesenteric lymph node, inguinal lymph node, tonsils, lungs, liver, kidneys, and heart) from ASFV-inoculated pigs (TRT pigs) euthanized at 3 dpi. A total 164 nodes and 2,444 interaction associations were detected. (B) Module analysis from the PPI network. The Top 3 significant modules (Module 1, 2, and 3) were obtained from the PPI network of DEGs, using MCODE. PPI, protein-protein interaction; ASFV, African swine fever virus; DEGs, differentially expressed genes; dpi, days post-inoculation.
Supplemental Material
Download JPEG Image (894.4 KB)Supplemental Material
Download JPEG Image (1 MB)Supplemental Material
Download JPEG Image (2.1 MB)Supplemental Material
Download JPEG Image (3.7 MB)Supplemental Material
Download JPEG Image (1.8 MB)Supplemental Material
Download JPEG Image (1.8 MB)Supplemental Material
Download JPEG Image (750.7 KB)Supplemental Material
Download MS Word (2.1 MB)Supplemental Material
Download MS Excel (4.8 MB)Supplemental Material
Download MS Excel (8.3 MB)Supplemental Material
Download MS Excel (1.4 MB)Supplemental Material
Download MS Excel (156 KB)Supplemental Material
Download MS Excel (1.2 MB)Supplemental Material
Download MS Excel (48.5 KB)Data availability statement
The transcriptomic datasets generated in this study can be found in the NCBI GEO database, under accession number 230340.
