Figures & data
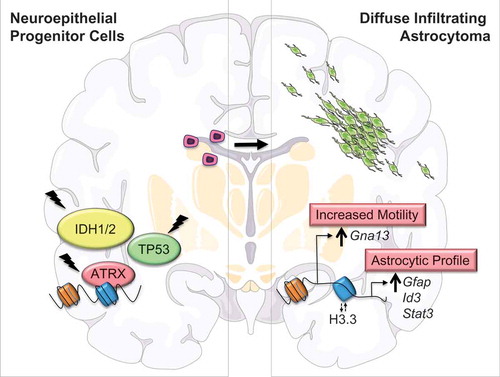
Figure 1. Epigenomic and transcriptional dysregulation occurring with ATRX deficiency drive disease-defining phenotypes in glioma cells of origin. ATRX (α-thalassemia mental retardation X-linked) loss of function mutations, together with IDH1/2 (isocitrate dehydrogenase enzymes 1 and 2) and TP53 (tumor protein p53) mutations, are defining molecular alterations characterizing the diffusely infiltrating astrocytomas. We demonstrated that Atrx inactivation alters chromatin structure and accessibility in the immediate vicinity of vacant Atrx binding sites (blue), in part due to shifts in the incorporation of the H3.3 histone variant. These changes induce the misexpression of locally situated genes, promoting the acquisition of disease-defining cellular phenotypes, such as motility and induction of astrocytic gene expression profiles.