Figures & data
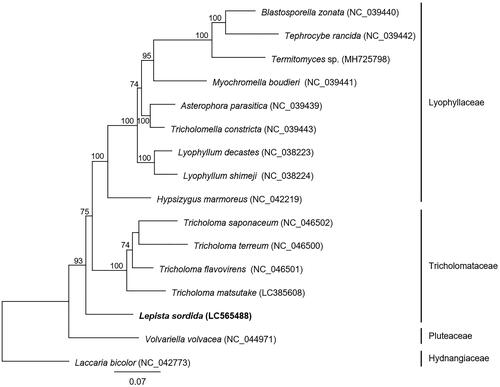
Figure 1. Molecular phylogenetic analysis of 16 basidiomycetes. The phylogenetic tree was constructed using the maximum-likelihood method with the amino acid sequences of 14 conserved mitochondrial proteins (cox1–3, cob, nad1–6, nad4L, atp6, atp8, and atp9). Laccaria bicolor was used as an outgroup species in the phylogenetic analysis. The accession numbers of the mitochondrial genome sequences used in this analysis are provided next to each species name. Bootstrap values higher than 70 are shown at the nodes. The scale bar indicates the number of substitutions per site.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov) under the accession number LC565488. The associated BioProject, SRA, and BioSample accession numbers are PRJDB4143, DRR327414, and SAMD00036984, respectively.
