Figures & data
Figure 1. The species reference image for Chrysojasminum subhumile. (A) A crow of C. subhumile; (B) a young branch of C. subhumile with yellow flowers and alternate leaves; the photos were taken by RenBin Zhu of Xishuangbanna Tropical Botanical Garden.

Figure 2. Graphic representation of features identified in Chrysojasminum subhumile chloroplast genome using cpgview (http://www.1kmpg.cn/cpgview). The map contains six rings. From the center outward, the first track shows the dispersed repeats. The dispersed repeats consist of direct (D) and Palindromic (P) repeats, connected with red and green arcs. The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors. The colors, the type of repeat they represent, and the description of the repeat types are as follows. Black: c (complex repeat); green: p1 (repeat unit size = 1); yellow: p2 (repeat unit size = 2); purple: p3 (repeat unit size = 3); blue: p4 (repeat unit size = 4); orange: p5 (repeat unit size = 5); red: p6 (repeat unit size = 6). The small single-copy (SSC), inverted repeat (IRa and IRb), and large single-copy (LSC) regions are shown on the fourth track. The GC content along the genome is plotted on the fifth track. The base frequency at each site along the genome will be shown between the fourth and fifth tracks. The genes are shown on the sixth track. The optional codon usage bias is displayed in the parenthesis after the gene name. Genes are color-coded by their functional classification. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively. The functional classification of the genes is shown in the bottom left corner.
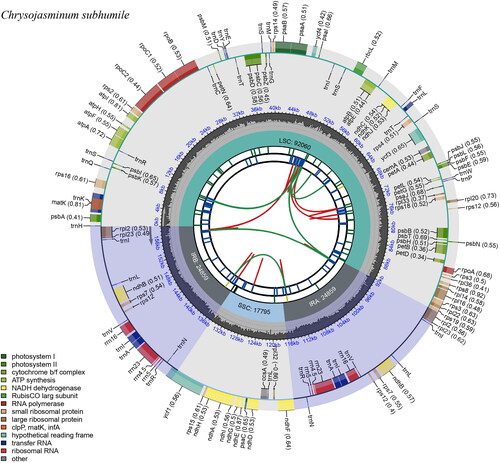
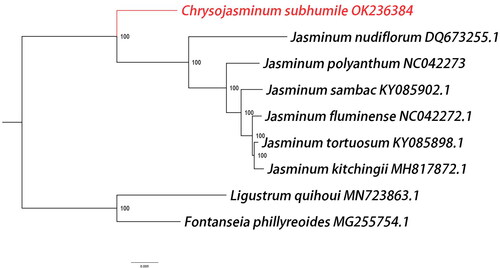
Figure 3. Phylogenetic relationships among nine complete chloroplast genomes of Oleaceae. The newly reported genome of Chrysojasminum subhumile was represented in red. Fontanesia phillyreoides and Ligustrum quihoui of Oleaceae family were taken as outgroups. Bootstrap support values are given at the nodes and the GenBank accession number was listed after the species name.
Supplemental Material
Download JPEG Image (259.8 KB)Supplemental Material
Download JPEG Image (82.5 KB)Supplemental Material
Download Text (3.7 KB)Supplemental Material
Download JPEG Image (1.4 MB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the number OK236384. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA842199, SRR19448979, and SAMN28649230, respectively.
