Figures & data
Figure 1. The image of Allogalathea elegans. Its thorax is extremely elongate, dorsally flattened, and ventrally carinate, with between five and nine lateral teeth, and a carapace covered in setiferous striae. The image A. elegans was taken by Ruan Xinhe on 25 December 2021.

Figure 2. The sequencing depth and coverage map for mitochondrial genomes. The average depth is 36.68×.
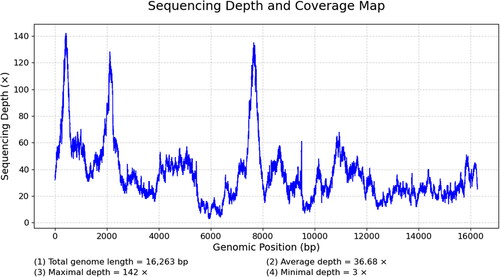
Figure 3. Mitogenome pattern map of Allogalathea elegans. The mitochondrial genome is mapped using CPGView. CDS: Coding sequence; tRNA: Transfer RNA; rRNA: Ribosomal RNA.
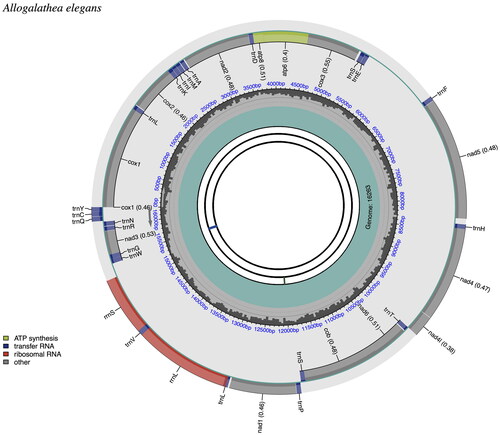
Table 1. The Organization of the complete mitochondrial genome in Allogalathea elegan.
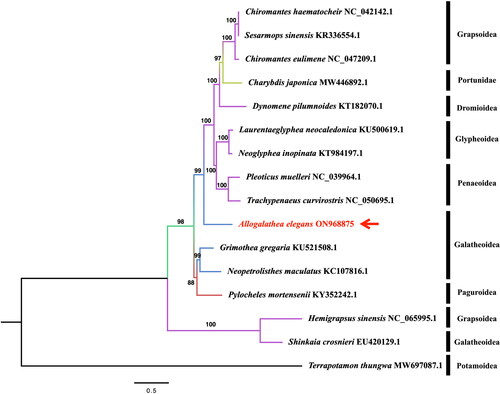
Figure 4. the phylogenetic tree was based on A. elegans and other 15 species, which was performed by ML analysis of the 13 protein-coding genes. The base composition and the phylogenetic tree were calculated using IQ-TREE 2.2.0 software with the maximum-likelihood method, 1000 replicates, and GTR + F+R4 model. The phylogenetic position of A. elegans was marked with a red arrow. The following sequences were used: Chiromantes haematochir NC_042142.1 (Li et al. Citation2019), Sesarmops sinensis KR336554.1 (Xing et al. Citation2016), Chiromantes eulimene NC_047209.1 (Zhang et al. Citation2020), Charybdis japonica MW446892.1 (Liu and Cui Citation2010), Dynomene pilumnoides KT182070.1 (Shi et al. Citation2016), Laurentaeglyphea neocaledonica KU500619.1 (Tan, Gan, Dally, et al. Citation2018), Neoglyphea inopinata KT984197.1 (Tan, Gan, Dally, et al. Citation2018), Pleoticus muelleri NC_039964.1 (Kim et al. Citation2018), Trachypenaeus curvirostris NC_050695.1 (Zhu et al. Citation2019), Grimothea gregaria KU521508.1 (Lee et al. Citation2016), Neopetrolisthes maculatus KC107816.1 (Shen et al. Citation2013), Pylocheles mortensenii KY352242.1 (Tan, Gan, Lee, et al. Citation2018), Hemigrapsus sinensis NC_065995.1 (Jin et al. Citation2023), Shinkaia crosnieri EU420129.1 (Yang and Yang Citation2008), Terrapotamon thungwa MW697087.1 (Yundaeng et al. Citation2022).
Data availability statement
The genome sequence data supporting this study’s findings are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under accession no. ON968875. The associated Bio-Project, SRA, and Bio-Sample numbers are PRJNA904942, SRR22403925, and SAMN31859346 respectively.
