Figures & data
Figure 2. Circular map of the assembled P. prolixa mitochondrial genome, consisting of 13 protein-coding, 22 transfer RNA, and two ribosomal RNA genes. Genes encoded on the reverse strand and forward strand are illustrated inside and outside the circles, respectively. This map was drawn using the OGDRAW web server (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html).
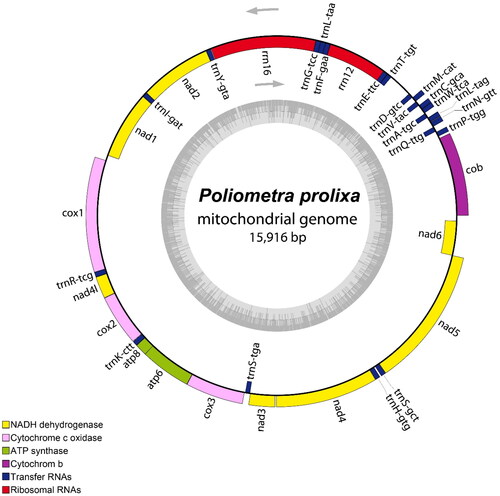
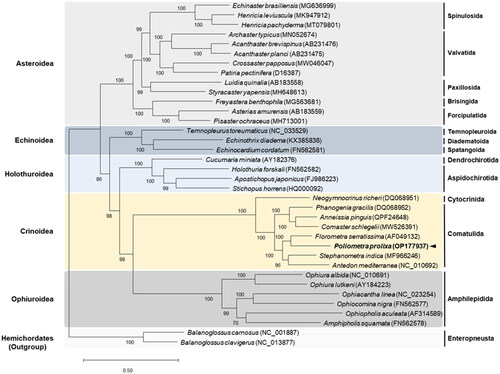
Figure 3. Maximum-Likelihood (ML) phylogenetic tree of 33 published complete Mitogenomes of Echinodermata, including that of P. prolixa, based on the concatenated nucleotide sequences of protein-coding genes (PCGs). The numbers on the branches indicate ML bootstrap percentages. DDBJ/EMBL/genbank accession numbers for published sequences are incorporated. The black arrow represents the sea pen analyzed in this study.
Supplemental Material
Download MS Word (627.6 KB)Data availability statement
BioProject, BioSample, and SRA accession numbers are https://www.ncbi.nlm.nih.gov/bioproject/PRJNA916601, https://www.ncbi.nlm.nih.gov/biosample/?term=SAMN32487547, and https://www.ncbi.nlm.nih.gov/sra/SRR22923304, respectively. The data that support the findings of this study are openly available in the National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov, with an accession number OP177937.

