Figures & data
Figure 1. The morphological traits of T. flavianalis, note the reddish-orange dorsal-fin and golden anal-fin (photographed by Fan Li).

Figure 2. Circular map of the assembled Tanichthys flavianalis mitogenome. Gene encoded on H- and L- strands with inverse arrow directions are shown outside and inside the circle, respectively. Plots of GC content and skew used a window size of 500 and reflect GC content/skew on a scale of 0 to 1 using a baseline of 0.5. Positive and negative skew are indicated by values above and below the midpoint respectively. This map was drawn using the Proksee web server.
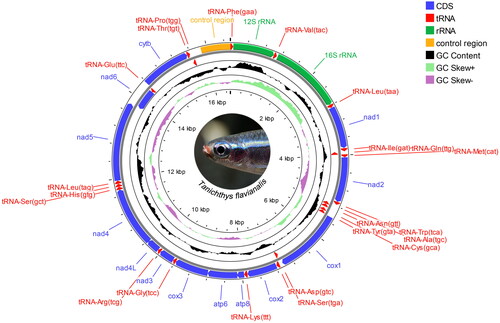
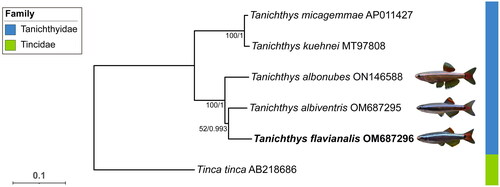
Figure 3. The phylogenetic tree constructed based on 13 protein coding genes of five Tanichthyidae mitogenomes and an outgroup. The support values of the corresponding nodes are shown (bootstrap values for ML/Bayes posterior probabilities for BI). The focal species is in bold, and pictures of the three Chinese Tanichthys were provided by Fan Li. The following sequences were used: T. micagemmae AP011427 (unpublished), T. kuehnei MT97808 (unpublished), T. albonubes ON146588 (unpublished), T. albiventris OM 687295 (Yang et al. Citation2023), T. flavianalis OM687296 (this study), and Tinca tinca AB218686 (Saitoh et al. Citation2006).
Supplemental Material
Download JPEG Image (54.7 KB)Supplemental Material
Download JPEG Image (110.9 KB)Supplemental Material
Download MS Word (13.2 KB)Supplemental Material
Download PDF (344 KB)Supplemental Material
Download MS Word (14.7 KB)Data availability statement
The genome sequence data of the study are deposited in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession no. OM687296. The associated BioProject, Bio-Sample and Run numbers are PRJNA957284, SAMN34248125, and SRR25113585, respectively.
