Figures & data
Figure 1. Pulsatilla chinensis f. alba D. K. Zang. Leaves 4 or 5, not fully expanded at anthesis; densely long pilose; leaf blade broadly ovate; 3-foliolate; margin entire or toothed. Sepals white. Wildlife photos were taken by C.B. in the Chinese city of Liaoyang, Liaoning Province. (E 123°33′09.26″, N 41°42′16.12″).

Figure 2. Chloroplast genome map of Pulsatilla chinensis f. alba D. K. Zang. From the center going outward, the first circle shows the forward and reverse repeats connected with red and green arcs, respectively. The second and third circles show the tandem repeats and microsatellite sequences marked with short bars, respectively. The outer circle shows the gene structure of the chloroplast genome. The genes were colored based on their functional categories, which were shown in the left corner. The map was drawn by cpgview (Liu et al. Citation2023).
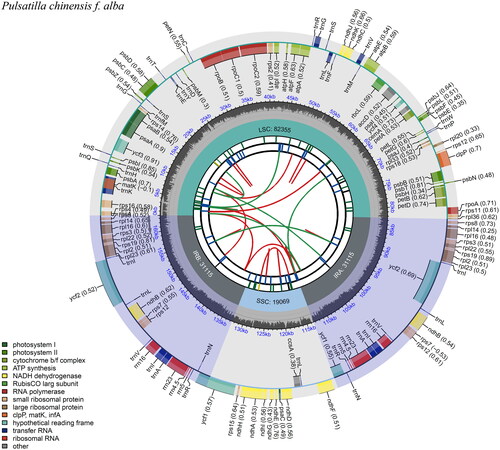
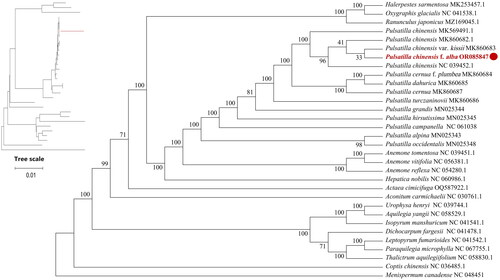
Figure 3. Maximum-likelihood (ML) phylogenetic tree of P. chinensis f. alba and 31 other complete chloroplast genome sequences. The numbers above the branches indicate the bootstrap values from ML analyses. The best evolutionary model was chosen as JTT + F+R2, which was selected using ModelFinder. The scale bar in the lower left corner of the figure represents the evolutionary distance, with a unit length of 0.01. The following sequences were used: Halerpestes sarmentosa (MK253457) (He et al. Citation2019), Oxygraphis glacialis NC_041538 (Zhai et al. Citation2019), Ranunculus japonicus MZ169045, P. chinensis MK569491, P. chinensis MK860682 (Zhang et al. Citation2019), Pulsatilla chinensis var. kissii MK860683 (Zhang et al. Citation2019), P. chinensis NC_039452 (Liu et al. Citation2018), P. cernua f. plumbea MK860684 (Zhang et al. Citation2019), P. dahurica MK860685 (Zhang et al. Citation2019), P. turczaninovii MK860686, P. cernua MK860687 (Zhang et al. Citation2019). P. grandis MN025344 (Li et al. Citation2020), P. hirsutissima MN025345 (Li et al. Citation2020), P. campanella NC_061038 (Xue et al. Citation2022), P. alpina MN025343 (Zhang et al. Citation2019), P. occidentalis MN025348 (Li et al. Citation2020). Anemone. tomentosa NC_039451 (Liu et al. Citation2018), A. vitifolia NC_056381, A. reflexa NC_054280 (Zhang et al. Citation2021), Hepatica nobilis NC_060986, Actaea cimicifuga OQ587922, Aconitum carmichaelii NC_030761, Urophysa henryi NC_039744, Aquilegia yangii NC_058529, Isopyrum manshuricum NC_041541 (Zhang et al. Citation2019), Dichocarpum fargesii NC_041478 (Zhang et al. Citation2019), Leptopyrum fumarioides NC_041542 (Zhang et al. Citation2019), Paraquilegia microphylla NC_067755, Thalictrum aquilegiifolium NC_058830, Coptis chinensis NC_036485, Menispermum canadense NC_048451.
Supplemental Material
Download MS Word (1 MB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under accession NO. OR085847. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA978389, SRX20587082 (Illumina), and SAMN35555254, respectively.
