Figures & data
Figure 1. Morphology and habitat map of Dendrocalamus liboensis Hsueh & D. Z. Li (These photographs were taken by Guangqian Gou in Libo, Guizhou, China). (A) Clump habit; (B) Leaf branch; (C) Internode. D. liboensis is an evergreen bamboo with 8–15 m culms, initially densely white powdery, branches usually from 6th or 7th node up, Central branch dominant and branchlets with 3–9 leaves.

Figure 2. Map of the D. liboensis chloroplast genome(GenBank: OR083041). This map was drawn using the OGDRAW. Genes inside the circle are transcribed clockwise, and those on the outside are transcribed counter-clockwise. Genes with related functions are shown in the same color. The darker gray in the inner circle corresponds to DNA G + C content, while the lighter gray corresponds to A + T content. LSC: large single-copy; SSC: small single-copy; IR: inverted repeat.
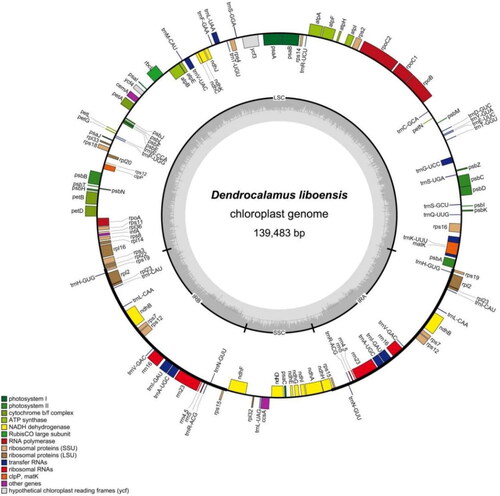
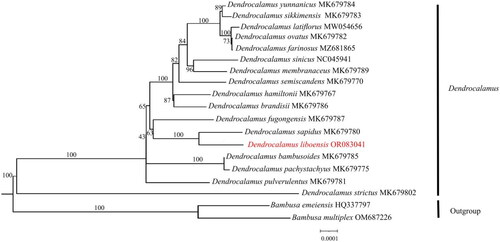
Figure 3. Maximum-likelihood(ML) phylogenetic tree of 19 species by IQtree based on complete chloroplast genomes, with Bambusa emeiensis HQ337797 (Zhang et al. Citation2011) and Bambusa multiplex OM687226 as outgroups. The phylogenetic tree was constructed using the maximum-likelihood method (ML) and bootstrap was performed 1000 times. Numbers on the nodes represent the bootstrap support values. The red fonts represents the assembled plastome sequence in this study. The following sequences were used: Dendrocalamus bambusoides MK679785 (Liu et al. Citation2020), Dendrocalamus birmanicus MK679772 (Liu et al. Citation2020), Dendrocalamus brandisii MK679786 (Liu et al. Citation2020), Dendrocalamus farinosus MZ681865, Dendrocalamus hamiltonii MK679767 (Liu et al. Citation2020), Dendrocalamus latiflorus MW054656(Liu et al. Citation2020), Dendrocalamus membranaceus MK679789 (Liu et al. Citation2020), Dendrocalamus fugongensis MK679787 (Liu et al. Citation2020), Dendrocalamus ovatus MK679782 (Liu et al. Citation2020), Dendrocalamus pachystachyus MK679775 (Liu et al. Citation2020), Dendrocalamus pulverulentus MK679781 (Liu et al. Citation2020), Dendrocalamus sapidus MK679780 (Liu et al. Citation2020), Dendrocalamus semiscandens MK679770 (Liu et al. Citation2020), Dendrocalamus sikkimensis MK679783 (Liu et al. Citation2020), Dendrocalamus sinicus NC045941 (Wang et al. Citation2019), Dendrocalamus strictus MK679802 (Liu et al. Citation2020), Dendrocalamus yunnanicus MK679784 (Liu et al. Citation2020). Undescribed citations in the legend indicate that the citations have not been published.
Supplemental Material
Download MS Word (171 KB)Data availability statement
Genome sequence data that support the findings of this study can be obtained from GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under accession no. OR083041. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA978368, SRR24793390, and SAMN35387695 respectively.
