Figures & data
Table I. Origin (country and location), GenBank or BOLD accession numbers and haplotype of the Gymnocephalus specimens analysed for molecular data
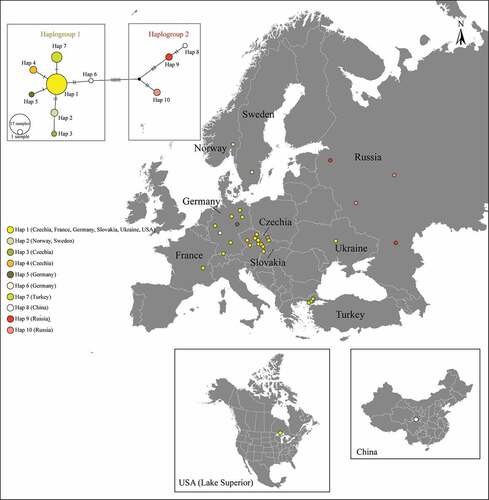
Figure 1. Neighbour-Joining (NJ) tree based on cytochrome c oxidase subunit I (COI) barcode gene sequences of ruffe Gymnocephalus cernua, Danube ruffe Gymnocephalus baloni, Gymnocephalus ambriaelacus and schraetzer (striped ruffe) Gymnocephalus schraetser, plus pike-perch Sander lucioperca as an outgroup. Bootstrap values (maximum likelihood-NJ) are shown above nodes if 50% or higher in value.
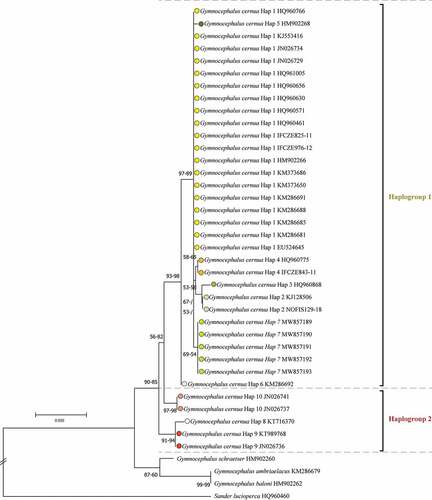
Table II. Scoring output (mean values from two assessors) from the Aquatic Species Invasiveness Screening Kit for ruffe Gymnocephalus cernua in the risk assessment areas of Thrace and Anatolia (Turkey)
Table III. Outcomes of the environmental impact classification of Alien Taxa (EICAT: Blackburn et al. Citation2014) applied to G. cernua in its introduced ranges of Europe and North America