Figures & data
Table I. The list of studied water bodies (lakes, ponds, reservoirs, rivers, brackish habitats) in Central Europe with basic morphometric and trophic characteristics. The trophic state was calculated based on Secchi disc visibility (hypertrophic <1 m; eutrophic 1–3 m; mesotrophic 3–5 m; oligotrophic >5 m). The number of lakes (no.) corresponds to the map with the distribution of three Diaphanosoma species in Central Europe (). Species: B - D. brachyurum, O - D. orghidani, M - D. mongolianum.
Figure 1. Distribution of three Diaphanosoma species (D. brachyurum, D. orghidani, D. mongolianum) in Central Europe. The dotted line shows the last glacial maximum.
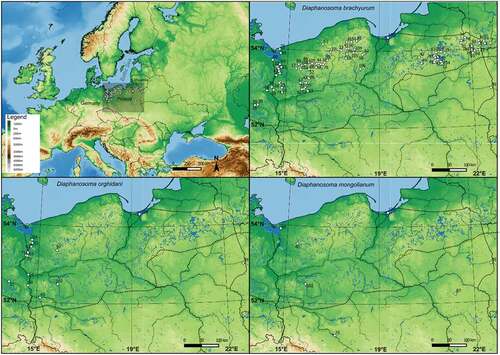
Figure 2. General view of the body and postabdominal claws Diaphanosoma brachyurum (a, b), D. mongolianum (c, d), and D. orghidani (e, f). Scale bars are 100 µm.
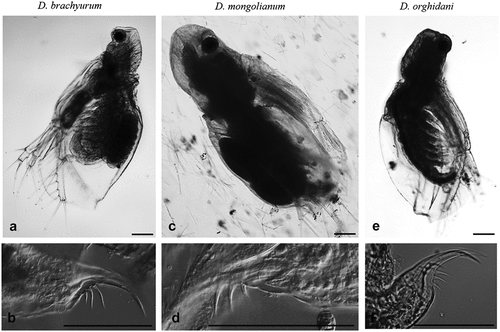
Figure 3. Armament of postero-ventral valve margins of D. brachyurum (a – Boczne; b – Kierzlińskie; c - Buwełno; d – Siemianówka), D. mongolianum (e, f - Siemianówka), and D. orghidani (g – Siemianówka; h - Oder River-warm canal). Scale bars are 50 µm.
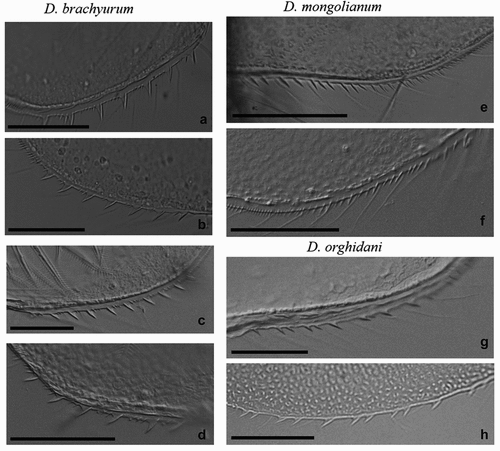
Figure 4. Body sizes of three Diaphanosoma species (a), and body sizes of D. brachyurum in different trophic conditions (b). The lower and upper limits of the boxes are the first and third quartiles, respectively. The crosses correspond to the means and the central horizontal bars are the medians. Points above or below the whiskers are the outliers and extreme values. The different letters (a, b) above the box plots denote significantly different values at p < 0.05; the same letters denote no statistically significant differences.
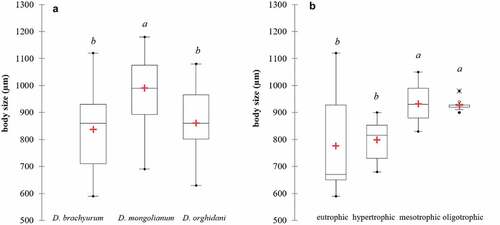
Figure 5. Diaphanosoma cf. lacustris from Vistula Lagoon. Specimen selected for the genetic analysis – haplotype H27. General view of the body (a), postabdominal claw (b), and armament of postero-ventral valve margins (c). Scale bars are 100 μm (a and b) and 50 μm (c).
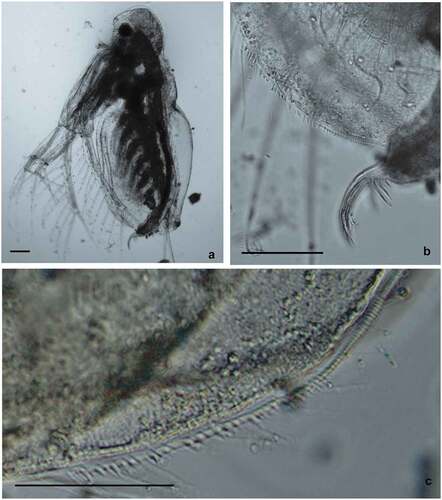
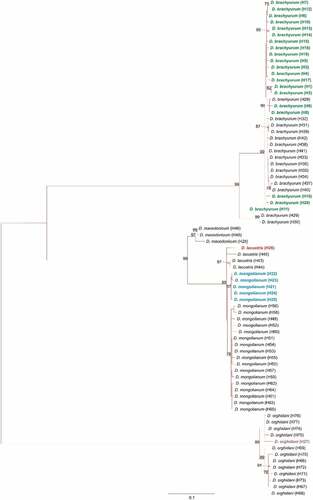
Figure 6. Maximum-likelihood tree computed with the GTR+I+G model of sequence evolution, representing phylogenetic relationships among the sequences of mitochondrial cytochrome c oxidase subunit I (COI) of Diaphanosoma species found in our study: D. brachyurum (H1 – H20, marked with green color); D. mongolianum (H21 – H25, marked with blue color); D. lacustris (H26 marked with red color); D. orghidani (H27 marked with a purple color) and downloaded (H28 – H77) from GenBank. Detailed information in Supplementary Information – Table S1. Numbers listed at nodes represent percent support for that node from 1,000 bootstrap replicates.