Figures & data
Table 1 Colonization, external hyphae density and spore production of arbuscular mycorrhizal fungi, as influenced by different trap plants
Figure 1 PCR-DGGE profiles of AM fungal communities recovered from the native soil (A), colonized roots (B∼H) and pot substrates (I∼O). Trap plants included sorghum (B, I), clover (C, J), maize (D, K), sorghum/clover (E, L), maize/sorghum (F, M), clover/maize (G, N), maize/clover/sorghum (H, O).
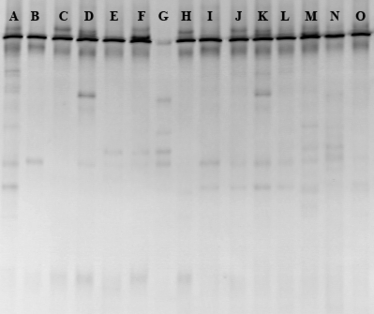
Table 2 Shannon–Weiner’s diversity index and species richness of indigenous arbuscular mycorrhizal fungi in the roots and pot media of different trap plants, as revealed by the band patterns in the polymerase chain reaction-denaturing gradient gel electrophoresis profile
Table 3 Sorenson’s pairwise similarity values between the native soil and the trap plant combinations on the basis of polymerase chain reaction-denaturing gradient gel electrophoresis profiles of arbuscular mycorrhizal fungi communities
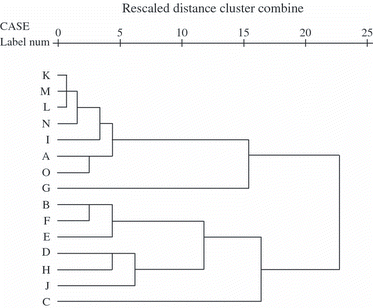
Figure 2 Dendrogram using average linkage (between groups) according to the Cs calculated on the basis of PCR-DGGE profiles of AM fungal communities in the native soil (A) and in colonized roots (B∼H) or pot substrate (I∼O) of different trap plants. Trap plants included sorghum (B, I), clover (C, J), maize (D, K), sorghum/clover (E, L), maize/sorghum (F, M), clover/maize (G, N), maize/clover/sorghum (H, O).