Figures & data
FIG 1 Epistasis analysis of Tel1 and Mrc1 mutants on G2/M cell cycle extension and cell viability. (A) ETI mrc1AQ rad9Δ synthetic phenotype shown via semiquantitative serial dilutions. ETI cells show reduced colony formation and colony size in the absence of functional Mrc1 and Rad9 proteins. Colony size and growth phenotypes are restored with deletion of Sml1. (B) Deletion of the Mrc1 partner protein, Tof1, does not mimic the ETI synthetic phenotype. Two biological replicates are shown.
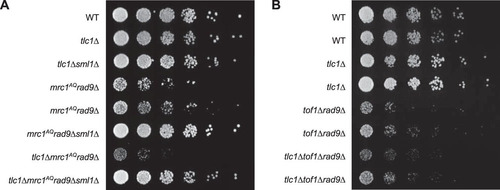
FIG 2 ETI, combined with loss of MRC1 and RAD9, leads to massive cell death without excessive telomere shortening. (A) Southern blot, using a Y′ telomere-specific probe to measure telomere length. (B) Serial streaks of freshly sporulated tlc1Δ strains starting approximately 20 generations after telomerase loss. The first, second, and third streaks represent cells after 40, 60, and 80 generations of telomerase loss, respectively. (C) FACS analysis of cells prepared from plates after serial streaks. Cultures were grown up, and samples were collected every 15 min following α-factor arrest and release. (D) WT and tlc1Δ mrc1AQ rad9 strains were freshly sporulated and dissected on YPD plates. Serial streaks were made from spore colonies, and 100 unbudded cells were identified by microscopic visualization and gridded individually onto YPD plates. The numbers of cells in descendant colonies were determined for each gridded cell after 16 and 37 h for the first two streaks (passage 1 is shown). Most cells ceased dividing as unbudded cells. Comparable wild-type cells yielded colonies so dense that cell numbers could not be determined. Red bars represent the first passage at 16 h, and green bars represent the second passage at 37 h.
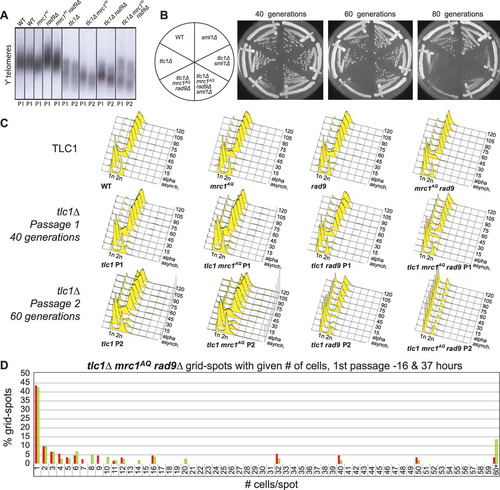
FIG 3 ETI mutants show dependence on the DNA damage response that can be alleviated by Sml1 deletion. Three different ETI mutants, the tlc1Δ (A), est2D530A (B), and est2Δ (C) mutants, showed loss of viability in strains lacking a functional DNA damage response due to Mrc1 and Rad9 mutation. Deletion of the RNR inhibitor, Sml1, alleviated this requirement (compare left and right halves of the plates). First-passage cells were taken directly from sporulation of diploid strains heterozygous for all the mutations. Three passages are shown (left to right), each representing approximately 20 generations.
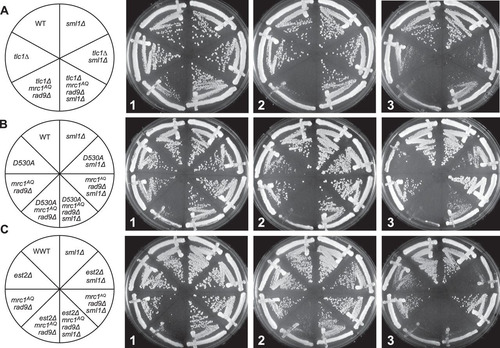
FIG 4 mrc1AQ rad9 cells arrest in G1 when telomeres lack telomerase. (B) In yEHB3284, a heterozygous diploid strain, telomeres were elongated through expression of Cdc13-Ku70p. After sporulation, mrc1AQ rad9 CDC13 cells with preelongated telomeres were selected and synchronized through α-factor arrest and release. Samples were collected before synchronization and every 15 min thereafter and analyzed by FACS.
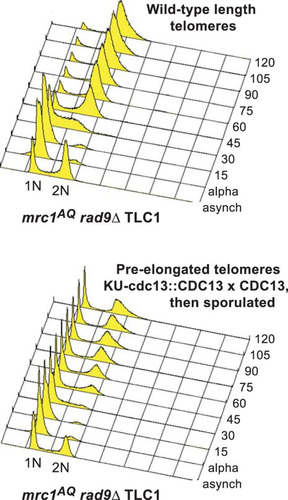
FIG 5 ETI cells do not display growth defects in the absence of Mec1 and Tel1 PIKK kinases. (A) sml1Δ, tel1Δ, and mec1Δ sml1Δ mutations had no effect on bulk colony growth phenotypes of ETI tlc1Δ strains (compare left and right halves of the plate). (B) ETI tlc1Δ mutation caused no additional growth defects in mec1 Δtel1Δ sml1Δ strains (compare left and right halves of the plates). (C) ETI est2Δ (as well as tlc1Δ and est2D530A [not shown]) caused no additional defects in mec1-21. All the streaks shown in panels A to C are first-passage cells taken directly from sporulation of diploid strains heterozygous for all the mutatons. (D) The ETI tlc1Δ mutation had no effect on the cell cycle profile of the mec1Δ and tel1Δ strains, regardless of telomere length. FACS analysis of synchronized WT, tel1Δ, mec1Δ sml1Δ, or tel1Δ mec1Δ sml1Δ cells was performed in the context of normal-length telomeres, ETI shortened telomeres, or telomeres lengthened through expression of and then selection against Cdc13-ku70p. Peaks represent DNA content: 1n at start and 2n at full duplication.
![FIG 5 ETI cells do not display growth defects in the absence of Mec1 and Tel1 PIKK kinases. (A) sml1Δ, tel1Δ, and mec1Δ sml1Δ mutations had no effect on bulk colony growth phenotypes of ETI tlc1Δ strains (compare left and right halves of the plate). (B) ETI tlc1Δ mutation caused no additional growth defects in mec1 Δtel1Δ sml1Δ strains (compare left and right halves of the plates). (C) ETI est2Δ (as well as tlc1Δ and est2D530A [not shown]) caused no additional defects in mec1-21. All the streaks shown in panels A to C are first-passage cells taken directly from sporulation of diploid strains heterozygous for all the mutatons. (D) The ETI tlc1Δ mutation had no effect on the cell cycle profile of the mec1Δ and tel1Δ strains, regardless of telomere length. FACS analysis of synchronized WT, tel1Δ, mec1Δ sml1Δ, or tel1Δ mec1Δ sml1Δ cells was performed in the context of normal-length telomeres, ETI shortened telomeres, or telomeres lengthened through expression of and then selection against Cdc13-ku70p. Peaks represent DNA content: 1n at start and 2n at full duplication.](/cms/asset/4000fe17-f967-4f3b-9178-e5dba1028a7f/tmcb_a_12276011_f0005.jpg)
FIG 6 Sml1 rescue is due to increased nucleotide pools. (A) Like sml1Δ, RNR1 overexpression (RNR1*) was able to rescue the colony formation defects of ETI mrc1AQ rad9Δ cells for all three ETI mutations. (B) Cells were serially diluted on rich-medium plates containing HU at the indicated concentrations. Despite a strong effect on control mec1Δ sml1Δ strains, no significant sensitivity to the drug was observed in ETI cells. Higher doses of the drug (25 mM and 100 mM) showed similar results (data not shown). (C) Dun1, the kinase responsible for phosphorylation and degradation of Sml1, was deleted. (D) Overexpression of Sml1 via a galactose-inducible promoter had no significant effect on growth in ETI strains relative to ETI alone. Frogged plates were incubated for 2 days (dextrose) or 3 days (galactose). (E) Elimination of recombination via Rad52 deletion did not eliminate the ETI mrc1AQ rad9Δ synthetic phenotype or prevent its rescue via Sml1 deletion. All the streaks shown are first-passage cells taken directly from sporulation of diploid strains heterozygous for all the mutations.
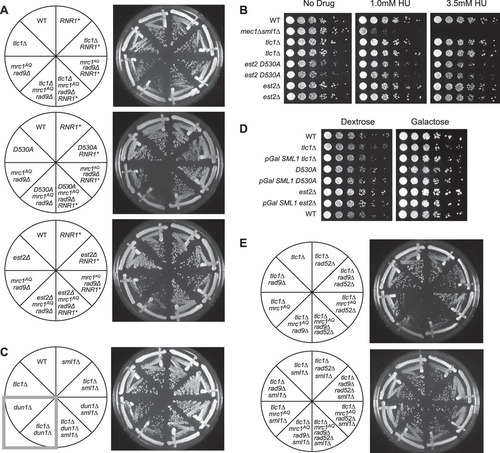
FIG 7 The ETI mrc1AQ rad9Δ synthetic phenotype and sm1Δ rescue cannot be explained by changing telomere lengths or accelerated senescence. Shown is Southern blot analysis of terminal XhoI restriction fragments. DNA was probed with an α-32P-labeled 5′-(TGTGGG)4-3′ Y′-specific probe. The lowest band represents the DNA fragment containing the terminal telomeric repeats. Telomere lengths for the first (1) and second (2) passages after sporulation of a diploid heterozygous strain are represented for all the genotypes. *, grid lines are added to reveal the inherent “smile” in the gel and to allow better comparison of lanes.
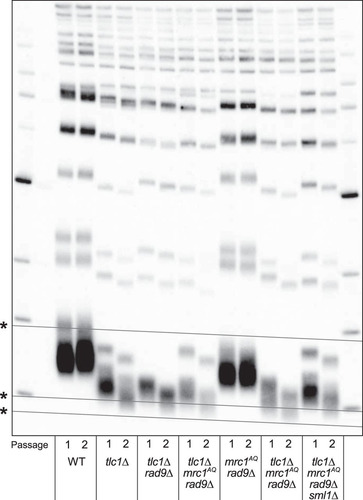
FIG 8 Hypomorphic telomerase does not show a synthetic phenotype with mrc1AQ rad9Δ mutation. (A) Southern blot analysis of terminal XhoI restriction fragments. DNA was probed with an α-32P-labeled 5′-(TGTGGG)4-3′ Y′ telomere-specific probe on samples from the indicated passages. (B and C) Streaks (B) and serial dilutions (C) of the hypomorphic TLC1 allele, tlc1-11. The tlc1-11 mutation did not create synthetic lethality with mrc1AQ rad9Δ mutations. In order to ensure shortened telomeres, all the tlc1-11 haploid strains were obtained by sporulation of a homozygous tlc1-11/tlc1-11 diploid, which was heterozygous for all the other mutations. (D) MRC1 is phosphorylated as telomeres become critically short. A Western blot of protein extracts was made from the same cultures used in panels A and E and probed with 9E10 anti-myc antibody. The control was WT protein extract treated with 200 mM hydroxyurea for 2 h. *, phosphorylation of Mrc1-myc13 after the indicated passages. (E) Cell cycle phase assignments made from FACS data on samples at 40 (P1) and 60 (P2) generations by plotting the DNA content (CellQuest Pro), followed by curve fitting and quantification of DNA peaks into G1, S, and G2/M categories (Flow-Jo).
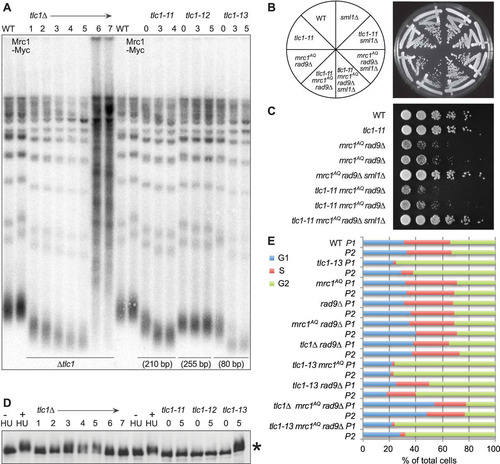
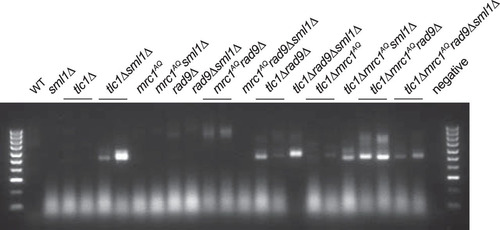
FIG 9 Rescue via Sml1 deletion may be partially due to reduction of deprotected telomeres in strains lacking a functional DNA damage response. A PCR assay was performed to detect deprotected telomeres that fused to an induced double-strand break (HO cut). The intensities of the bands represent the approximate numbers of captured deprotected telomeres. No DNA bands were visible in experiments when either primer was excluded from the PCR. Sml1 deletion had no effect or increased the number of deprotected telomeres detected in strains with a fully or partially functional DDR and reduced the number of deprotected telomeres detected in strains completely lacking a DDR (mrc1AQ rad9Δ).