Figures & data
TABLE 1 Saccharomyces cerevisiae strains used in this study
FIG 1 Localization of Gly1007 in the C128 subunit sequence and Pol III structure. (A) The C128 protein structure was modeled based on the respective RNA Pol I subunit A135. In the available 2.8-Å structure of RNA Pol I (PDB ID 4C2M), subunit A135 was replaced by the model of C128. The localization of selected subunits in the complex and position of Gly1007 of C128 are shown. This glycine (marked in red) is located in the C128 structure close to another conserved motif, DMPF (marked in blue), as revealed by magnification of the respective fragment of the polymerase structure in the wild-type (wt) (Gly1007 marked in red) and mutant (rpc128-1007) (Ala1007 marked in red) strains. The structure model was produced using the PyMOL program. (B) the DMPF motif (aa 929 to 932) and glycine 1007 in the C128 subunit of Pol III in S. cerevisiae (RPC128_S.c) are conserved in sequences of the second-largest subunits of yeast Pol I (RPA135_S.c) and Pol II (RPB2_S.c.), as well as in their human homologues (RPC2_H.s, RPA2_H.s, and RPB2_H.s, respectively).
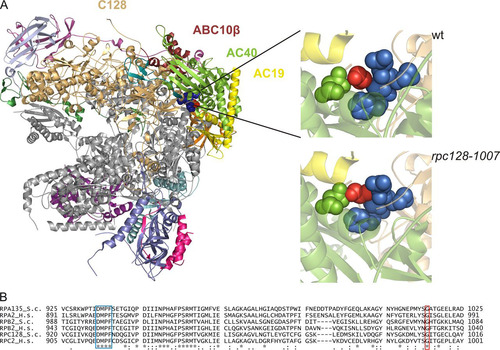
FIG 2 Inhibited growth and reduced levels of the largest subunit of Pol III in the rpc128-1007 mutant. (A) The rpc128-1007 mutant (MJ15-9C) and the control wt strain (MB159-4D) were inoculated (1/100) into liquid rich glucose medium (YPD) and incubated at 30°C with shaking. The growth was monitored by OD600 measurements. (B and C) rpc128-1007 cells encoding C160-HA were examined by immunofluorescence and Western blotting by using anti-HA antibody and anti-Rpb1 antibody. (D) As indicated, the rpc128-1007 mutant was transformed with empty vector [-], a plasmid harboring RPC128, or a plasmid harboring RPC160. Tenfold serial dilutions of overnight cultures in minimal medium (SC−ura) were plated on SC−ura and incubated at 30°C for 3 days or on rich glucose medium (YPD) and incubated at 16°C for 4 days.
![FIG 2 Inhibited growth and reduced levels of the largest subunit of Pol III in the rpc128-1007 mutant. (A) The rpc128-1007 mutant (MJ15-9C) and the control wt strain (MB159-4D) were inoculated (1/100) into liquid rich glucose medium (YPD) and incubated at 30°C with shaking. The growth was monitored by OD600 measurements. (B and C) rpc128-1007 cells encoding C160-HA were examined by immunofluorescence and Western blotting by using anti-HA antibody and anti-Rpb1 antibody. (D) As indicated, the rpc128-1007 mutant was transformed with empty vector [-], a plasmid harboring RPC128, or a plasmid harboring RPC160. Tenfold serial dilutions of overnight cultures in minimal medium (SC−ura) were plated on SC−ura and incubated at 30°C for 3 days or on rich glucose medium (YPD) and incubated at 16°C for 4 days.](/cms/asset/8b16c0e7-8096-4f66-ac36-e7576e81640e/tmcb_a_12275564_f0002_oc.jpg)
TABLE 2 Overdose suppressors of the rpc128-1007 mutation
FIG 3 ABC10β and Rbs1 as overdose suppressors of the rpc128-1007 mutation. As indicated, the rpc128-1007 mutant (MJ15-9C) and the control wt strain (MB159-4D) were transformed with empty vector [-], a plasmid harboring RBS1, or a plasmid harboring RPB10. (A) Tenfold serial dilutions of overnight cultures were plated on YPD following incubation at 30°C for 3 days or at 16°C for 4 days. For control of plasmid maintenance, the same cells were plated on minimal glucose medium (SC−ura) and incubated at 30°C for 3 days. (B) Cells preincubated in minimal medium were grown in YPD at 30°C, shifted to 16°C for 2 h, and harvested. Total RNA was isolated, separated on a 2.8% agarose gel, and stained with EtBr using equal amounts of RNA per lane (5 μg). (C) RNA was analyzed by Northern blotting using probes specific for pre-tRNA-Leu (CAA), tRNA-Phe (GAA), and tRNA-Tyr (GUA). 5.8S rRNA served as the loading control. Positions of initial transcripts, end-processed intron-containing pre-tRNA (IVS), and mature tRNA are indicated. (D) pre-tRNA forms were quantified. Bars represent levels for primary transcripts and intron-containing precursors normalized to the loading control. Standard deviations were estimated on the basis of three independent experiments. The P values calculated for ratios of pre-tRNAs (rpc128-1007[-]/wt[-], rpc128-1007[-]/rpc128-1007[RPB10], and rpc128-1007[-]/rpc128-1007[RBS1]) showed statistical significance (P < 0.05).
![FIG 3 ABC10β and Rbs1 as overdose suppressors of the rpc128-1007 mutation. As indicated, the rpc128-1007 mutant (MJ15-9C) and the control wt strain (MB159-4D) were transformed with empty vector [-], a plasmid harboring RBS1, or a plasmid harboring RPB10. (A) Tenfold serial dilutions of overnight cultures were plated on YPD following incubation at 30°C for 3 days or at 16°C for 4 days. For control of plasmid maintenance, the same cells were plated on minimal glucose medium (SC−ura) and incubated at 30°C for 3 days. (B) Cells preincubated in minimal medium were grown in YPD at 30°C, shifted to 16°C for 2 h, and harvested. Total RNA was isolated, separated on a 2.8% agarose gel, and stained with EtBr using equal amounts of RNA per lane (5 μg). (C) RNA was analyzed by Northern blotting using probes specific for pre-tRNA-Leu (CAA), tRNA-Phe (GAA), and tRNA-Tyr (GUA). 5.8S rRNA served as the loading control. Positions of initial transcripts, end-processed intron-containing pre-tRNA (IVS), and mature tRNA are indicated. (D) pre-tRNA forms were quantified. Bars represent levels for primary transcripts and intron-containing precursors normalized to the loading control. Standard deviations were estimated on the basis of three independent experiments. The P values calculated for ratios of pre-tRNAs (rpc128-1007[-]/wt[-], rpc128-1007[-]/rpc128-1007[RPB10], and rpc128-1007[-]/rpc128-1007[RBS1]) showed statistical significance (P < 0.05).](/cms/asset/6f17166d-2aba-4520-877e-573e9bf68498/tmcb_a_12275564_f0003_oc.jpg)
TABLE 3 Specificity of RBS1 and RPB10 suppressorsTable Footnotea
FIG 4 Synthetic negative growth defect of the double rpc128-1007 rbs1Δ mutant. Sporulation of diploids generated by the cross MB159-2C rbs1Δ × MJ15-9C rpc128-1007 was followed by dissection of tetrads. The growth of spores was monitored after 3 days and 5 days (left panel). Genetic analysis of all tetrads was performed by transferring all individual spores a to YPD master plate which was grown for 2 days and replicated to test the genetic phenotypes of Geneticin resistance and cold sensitivity (growth at 16°C). Genetic analysis of one selected tetrad is presented in the right panel.
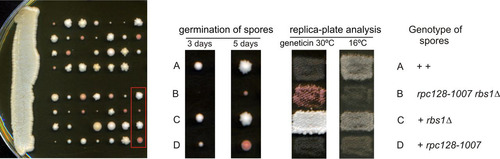
TABLE 4 Components of RNA polymerases copurified with Rbs1Table Footnotea
FIG 5 Rbs1 interacts with AC40. (A) Strain MJ15-10A, expressing Myc-tagged Rbs1, and control strain BY4741 rbs1Δ were grown in YPD. Aliquots of cell extracts (input) and immunoprecipitates with the anti-Myc antibody were analyzed by immunoblotting with anti-Myc and anti-AC40 antibodies. The lane in the middle of the gel is empty and separates input from IP with anti-Myc. (B) Cell extracts prepared from strains encoding GFP-tagged Rbs1, Maf1, and Rex1 proteins and control strain BY4741 rbs1Δ (input) and immunoprecipitates with the anti-GFP antibody were analyzed by immunoblotting with anti-GFP and anti-AC40 antibodies.
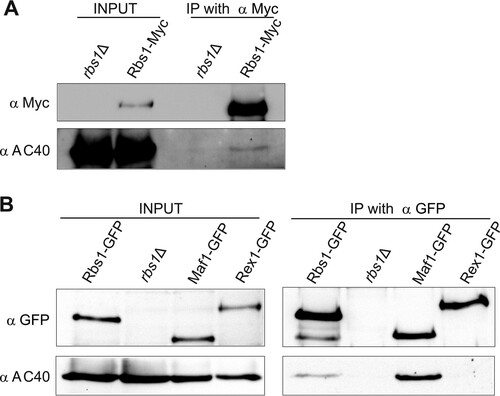
FIG 6 LMB induces nuclear accumulation of Rbs1. Yeast cells were grown to log phase at 30°C (−LMB), and then LMB (100 ng/ml) was added and growth was continued at 30°C for the indicated times before fixation of cells. Localization of Rbs1-Myc was determined by immunofluorescence with anti-Myc antibody in the control wild-type (wt) strain (A), the crm1-T359C mutant (B), and the crm1-T359C rpc128-1007 double mutant (C). Nuclear DNA was stained with DAPI (4′,6′-diamidino-2-phenylindole). (D) The percentage of cells with the given cellular localization of Rbs1 after 1 h of LMB treatment was estimated after inspection of at least 120 single cells.
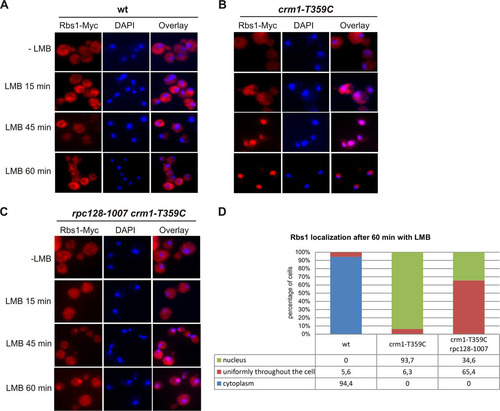
FIG 7 The rpc128-1007 mutation lowers the nuclear concentration of some Pol III subunits, but these effects are suppressed by overproduced Rbs1. Wild-type and rpc128-1007 mutant derivatives transformed with empty vector ([-]) or a multicopy plasmid with the RBS1 gene were grown in rich glucose medium to log phase. (A) Immunofluorescence of cells encoding C160-HA by using anti-HA antibody. Nuclei were stained with DAPI. (B) Live imaging of cells encoding C82-GFP or C53-GFP.
![FIG 7 The rpc128-1007 mutation lowers the nuclear concentration of some Pol III subunits, but these effects are suppressed by overproduced Rbs1. Wild-type and rpc128-1007 mutant derivatives transformed with empty vector ([-]) or a multicopy plasmid with the RBS1 gene were grown in rich glucose medium to log phase. (A) Immunofluorescence of cells encoding C160-HA by using anti-HA antibody. Nuclei were stained with DAPI. (B) Live imaging of cells encoding C82-GFP or C53-GFP.](/cms/asset/030e4640-4684-4edf-a2cc-439ac97522c0/tmcb_a_12275564_f0007_oc.jpg)
FIG 8 The defect of Pol III assembly in the rpc128-1007 mutant is partially rescued by overproduced Rbs1. Extracts were prepared from control wild-type cells and cells encoding C160-HA (wild-type and rpc128-1007 mutant) transformed with empty vector ([-]) or a multicopy plasmid with the RBS1 gene (input). Pol III was immunoprecipitated from yeast extracts by HA-specific antibody (IP with α HA). (A) Primary extracts (lanes 1 to 4) and IP fractions (lanes 6 to 9) were examined by Western blotting with antibody specific for HA, C82, C53, AC40, or Vma2 (used as a loading control). An additional higher band which was repeatedly recognized by anti-C82 antibody in mutant cells appeared to be a result of an unspecific cross-reaction. Lane 5 is empty and separates input from IP with anti-HA. (B) Quantification of the levels of Pol III subunits in total extracts from the rpc128-1007 mutant transformed with empty vector or a multicopy plasmid with the RBS1 gene. Band intensities from Western blot images were quantified by MultiGauge v3.0 software (Fujifilm). Bars represent the means with standard deviations from three independent experiments. Levels from the wild-type strain were assumed to be 1. (C) Quantification of the amount of Pol III subunits coimmunopurified with C160 from extracts of the rpc128-1007 mutant transformed with empty vector or a multicopy plasmid with the RBS1 gene. Bands intensities from Western blot images were calculated as for panel B. The subunit ratios in the wild-type, rpc128-1007 [-], and rpc128-1007 [RBS1] strains were calculated and referred to levels of the respective subunits in the wild-type strain, which were assumed to be 1.
![FIG 8 The defect of Pol III assembly in the rpc128-1007 mutant is partially rescued by overproduced Rbs1. Extracts were prepared from control wild-type cells and cells encoding C160-HA (wild-type and rpc128-1007 mutant) transformed with empty vector ([-]) or a multicopy plasmid with the RBS1 gene (input). Pol III was immunoprecipitated from yeast extracts by HA-specific antibody (IP with α HA). (A) Primary extracts (lanes 1 to 4) and IP fractions (lanes 6 to 9) were examined by Western blotting with antibody specific for HA, C82, C53, AC40, or Vma2 (used as a loading control). An additional higher band which was repeatedly recognized by anti-C82 antibody in mutant cells appeared to be a result of an unspecific cross-reaction. Lane 5 is empty and separates input from IP with anti-HA. (B) Quantification of the levels of Pol III subunits in total extracts from the rpc128-1007 mutant transformed with empty vector or a multicopy plasmid with the RBS1 gene. Band intensities from Western blot images were quantified by MultiGauge v3.0 software (Fujifilm). Bars represent the means with standard deviations from three independent experiments. Levels from the wild-type strain were assumed to be 1. (C) Quantification of the amount of Pol III subunits coimmunopurified with C160 from extracts of the rpc128-1007 mutant transformed with empty vector or a multicopy plasmid with the RBS1 gene. Bands intensities from Western blot images were calculated as for panel B. The subunit ratios in the wild-type, rpc128-1007 [-], and rpc128-1007 [RBS1] strains were calculated and referred to levels of the respective subunits in the wild-type strain, which were assumed to be 1.](/cms/asset/8eee961f-000f-4bb2-9f9e-14f1ee6d1f5d/tmcb_a_12275564_f0008_oc.jpg)
