Figures & data
Figure 1 Overview of a set of bioinformatics and visual analytics methods used to prioritize protein sequences for further research.
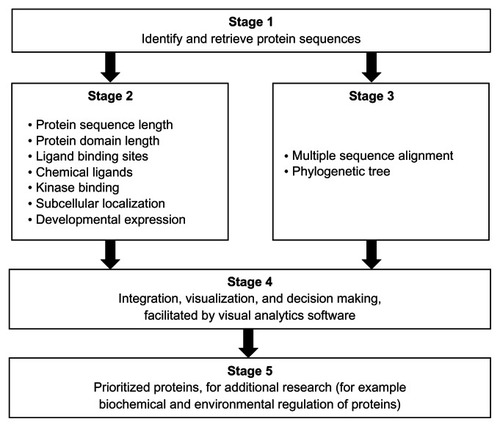
Table 1 Annotation features for universal stress proteins of Schistosoma mansoni and Schistosoma japonicum
Figure 2 Grouping of 13 Schistosoma USPs by sequence length.
Abbreviations: aa, amino acid; UniProt, Universal Protein Resource (Apweiler et al);Citation47 USP, universal stress protein.
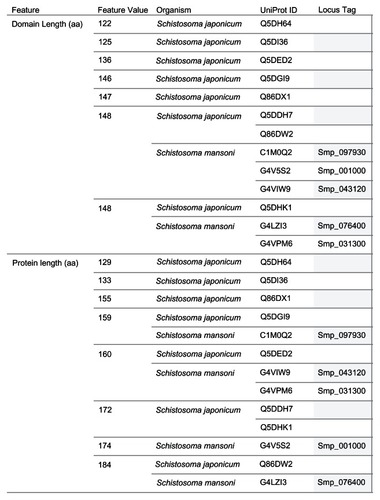
Figure 3 Grouping of 13 Schistosoma universal stress proteins by functional site signature.
Abbreviations: ATP, adenosine triphosphate; UniProt, Universal Protein Resource; USP, universal stress protein.
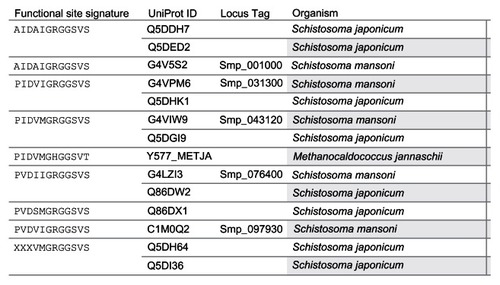
Figure 4 Multiple sequence alignment of the sequences of selected universal stress proteins of Schistosoma mansoni and Schistosoma japonicum.
Abbreviations: ATP, adenosine triphosphate; D, aspartate; G, glycine; H, histidine; L, leucine; P, proline; Uniprot, Universal Protein Resource; USP, universal stress protein.
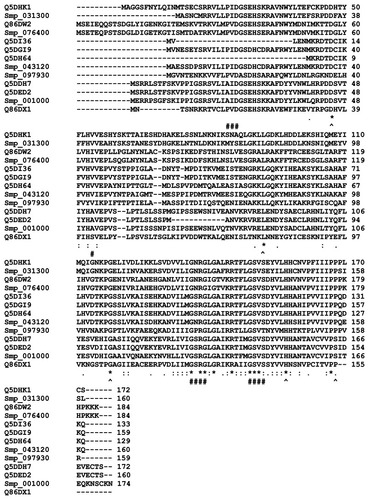
Figure 5 Grouping of 13 Schistosoma universal stress protein sequences. The phylogenetic tree was generated with MEGA5 (Tamura et al),Citation59 using the maximum likelihood method. The 13 Schistosoma universal stress protein sequences were clustered in five groups (A–E). The numbers near the clades are the statistics from the 1000 bootstrap that support the phylogeny recovery of the clades. A visual analytics resource that can be used to view the image, with other associated data, is available at http://public.tableausoftware.com/views/schisto_features_usp/phylotrees. Sequences of S. mansoni have “Smp” in the sequence identifier.
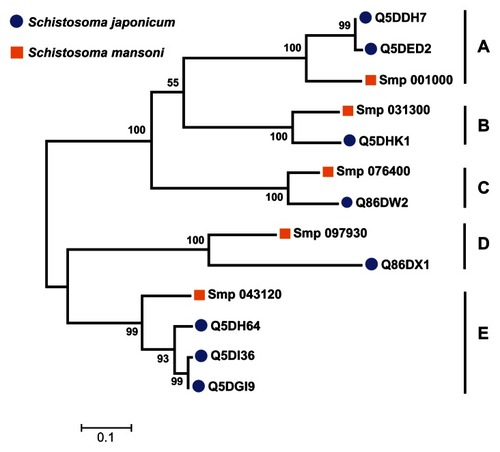
Figure 6 Parsimony test for the phylogeny tree reconstructed for Schistosoma universal stress proteins with the maximum likelihood method.
Figure 7 Integration and visualization of the data on the sequence features, evolutionary relatedness, and developmental expression of Schistosoma universal stress proteins (Q86DW2 and G4LZI3).
Abbreviations: ADP, adenosine diphosphate; AMP, adenosine monophosphate; ATP, adenosine triphosphate; CA, calcium; D, aspartate; G, glycine; GTP, guanosine triphosphate; I, isoleucine; Mg, magnesium; PKC, protein kinase C; P, proline; R, arginine; S, serine; UniProt, Universal Protein Resource (Apweiler et al);Citation47 V, valine; Zn, Zinc.
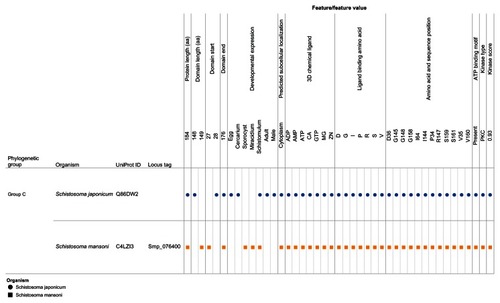
Figure 8 Integration and visualization of the data on the sequence features, evolutionary relatedness, and developmental expression of Schistosoma universal stress proteins (Q86DX1 and C1M0Q2).
Abbreviations: ADP, adenosine diphosphate; AMP, adenosine monophosphate; ATP, adenosine triphosphate; D, aspartate; G, glycine; GTP, guanosine triphosphate; I, isoleucine; M, methionine; Mg, magnesium; PKA, protein kinase A; PKC, protein kinase C; P, proline; R, arginine; S, serine; T, threonine; UniProt, Universal Protein Resource (Apweiler et al);Citation47 V, valine; Zn, Zinc.
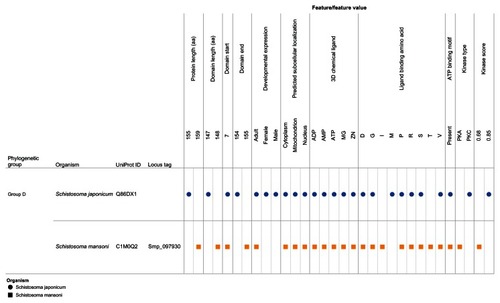
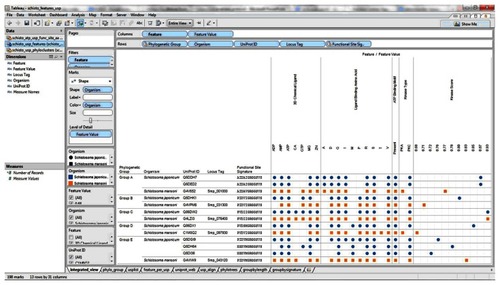
Figure 9 Design layout and visualization of data sets from the sequence analysis, evolutionary relatedness, and developmental expression of 13 Schistosoma universal stress proteins.
Abbreviations: ADP, adenosine diphosphate; AMP, adenosine monophosphate; ATP, adenosine triphosphate; CA, calcium; D, aspartate; G, glycine; GTP, guanosine triphosphate; I, isoleucine; M, methionine; Mg, Magnesium; PKA, protein kinase A; PKC, protein kinase C; P, proline; R, arginine; S, serine; T, threonine; UniProt, Universal Protein Resource (Apweiler et al);Citation47 V, valine; Zn, Zinc.