Figures & data
Table 1 Associations Between TRIM58 Expression Level and Baseline Clinicopathological Characteristics in Breast Cancer
Figure 1 Associations between TRIM58 expression and clinicopathological characteristics of all patients. Associations between TRIM58 expression and tumor size (A), molecular subtype (B), ER status (C), PR status (D), Ki67 index (E), and LN status (F). The P values are calculated using the Mann–Whitney U-test.
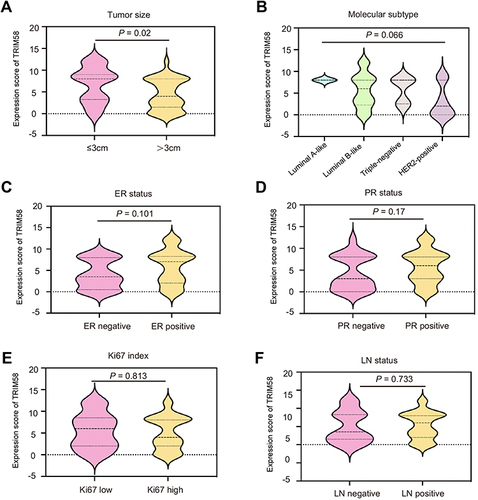
Figure 2 Clinicopathological features of pathological complete response (pCR; n = 21) and no-pCR (n = 37) patients. (A) Two-category data (TRIM58 expression high versus low, age ≥50 versus <50 years, tumor size ≥ 3 versus < 3, LN status positive versus negative, ER positive versus negative, PR positive versus negative, HER2 positive versus negative, and Ki67 index ≥30% versus <30%) are shown with values 1 and 0, respectively.
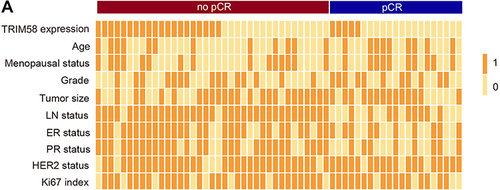
Table 2 Univariate and Multivariate Analyses for the Predictive Factors of Pathological Complete Response
Figure 3 Different TRIM58 immunochemistry staining. (A). Weak TRIM58 expression with a staining score of 1 (≤ 6). (B). Weak TRIM58 expression with a staining score of 6 (≤ 6). (C). Strong TRIM58 expression with a staining score of 8 (> 6). (D). Strong TRIM58 expression with a staining score of 12 (> 6).
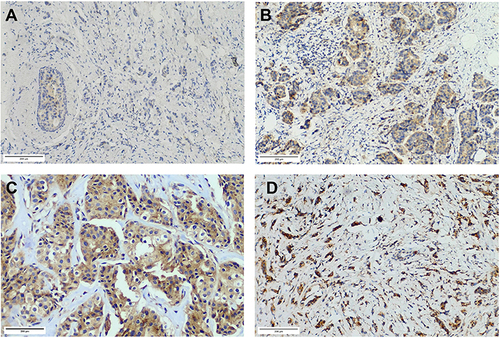
Figure 4 Receiver operating characteristic curves of different predictive models for pathological complete response. (A) The blue line exhibits model 1 (AUC 0.885, incorporating TRIM58 expression, age, menopausal status, T stage, N stage, ER status, PR status, HER2 status, Ki67 index and histological grade). The green line shows model 2 (AUC 0.844, including all the factors in model 1 except TRIM58 expression). The red line represents model 3 (AUC 0.778, integrating TRIM58 expression and AJCC stage). The pink line is presented with model 4 (AUC 0.719, AJCC stage alone). The AUC values are compared using the z-test.
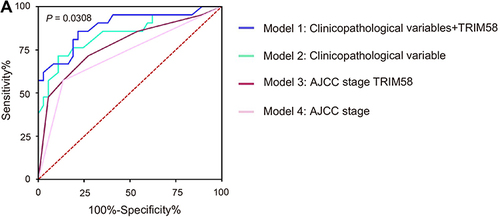
Figure 5 Kaplan–Meier survival curves according to TRIM58 expression in Kaplan-Meier Plotter datasets. Kaplan–Meier estimates of overall survival (OS) for all patients (A), HorR+ subgroup (B), HorR- subgroup (C), HER2+ subgroup (D) and HER2- subgroup (E). The P values are calculated using the Log rank test. Hazard ratios are derived from the Cox proportional hazards regression model.
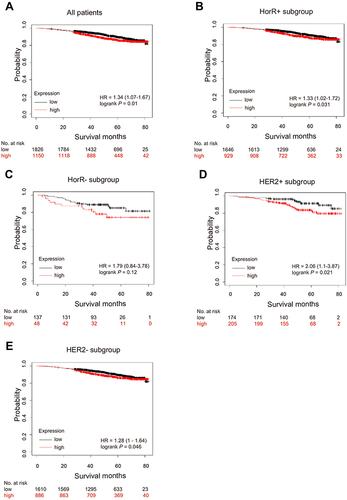
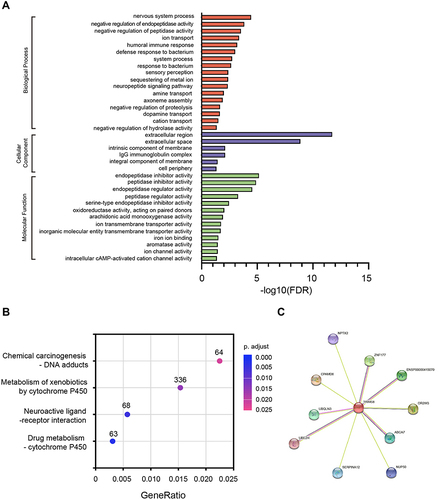
Figure 6 Gene ontology categories, Kyoto Encyclopedia of Genes and Genomes pathways, and protein–protein interaction network associated with TRIM58. Gene ontology categories (A); Kyoto Encyclopedia of Genes and Genomes pathways (B); and protein–protein interaction network (C) associated with TRIM58.