Figures & data
Table 1 Characteristics of studies included for the association between P16INK4a methylation and ovarian cancer risk
Table 2 Characteristics of studies included for the association between P16INK4a methylation and clinicopathological features of ovarian cancer
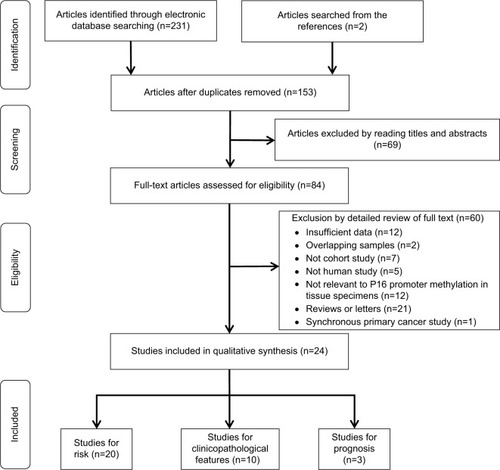
Figure 2 Forest plots for the association between P16INK4a methylation and ovarian cancer risk.
Notes: (A) Cancer tissues vs normal tissues; (B) cancer tissues vs benign tissues; (C) cancer tissues vs LMP tissues.
Abbreviations: LMP, low malignant potential or borderline tumor tissues; M–H, Mantel–Haenszel.
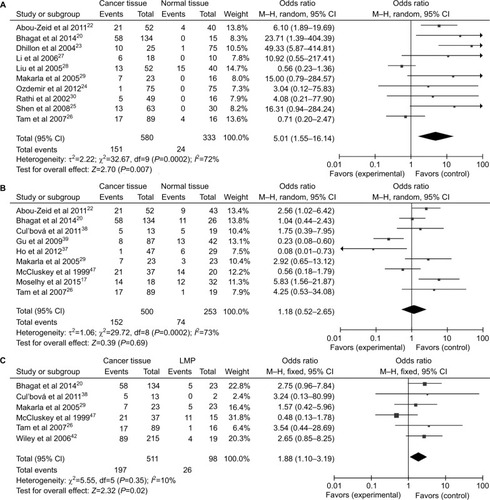
Figure 3 Forest plots for the association between P16INK4a methylation and ovarian diseases.
Notes: (A) Benign tissues vs normal tissues; (B) LMP tissues vs normal tissues.
Abbreviations: LMP, low malignant potential or borderline tumor tissues; M–H, Mantel–Haenszel.
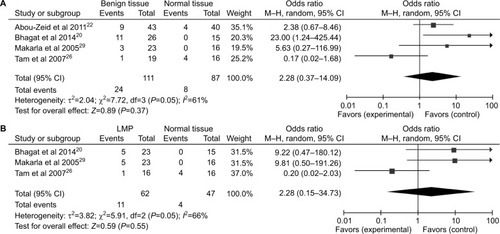
Table 3 Meta-regression and subgroup analyses of P16INK4a methylation in comparison of cancer tissues vs normal tissues
Figure 4 Forest plots for the association between P16INK4a methylation and clinicopathological features in ovarian cancer.
Notes: (A) Age; (B) clinical stage; (C) tumor grade; (D) histological subtype.
Abbreviation: M–H, Mantel–Haenszel.
Figure 5 Forest plots for P16INK4a methylation on survival analysis in univariate and multivariate Cox regression model.
Notes: (A) PFS in univariate Cox regression model; (B) PFS in multivariate Cox regression model; (C) OS in univariate Cox regression model; (D) OS in multivariate Cox regression model.
Abbreviations: OS, overall survival; PFS, progression-free survival; SE, standard error.
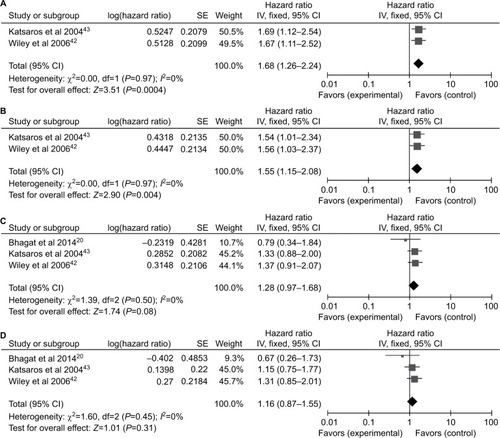
Figure 6 Sensitivity analyses and Begg’s test for publication bias of P16INK4a methylation during the carcinogenesis of ovarian cancer.
Notes: (A and D) Cancer tissues vs normal tissues; (B and E) sensitivity analysis for the comparison of cancer tissues vs benign tissues; (C and F) sensitivity analysis for the comparison of cancer tissues vs LMP tissues.
Abbreviations: LMP, low malignant potential or borderline tumor tissues; SE, standard error.
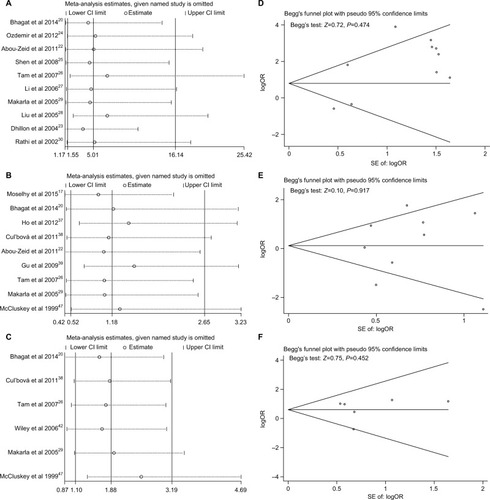
Table 4 Methylation of P16INK4a CpG sites on Illumina HumanMethylation 27 BeadChip from TCGA datasets