Figures & data
Figure 1 The genetic alteration of the genes involved in the E-cadherin regulation process. We proceeded to the TCGA database and finally identified 26 genes which were frequently altered (>2%) in HCC.
Notes: This figure is from the cBioPortal website (http://www.cbioportal.org/); TCGA data set. Cerami E, Gao J, Dogrusoz U, et al The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404.Citation51 Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1.Citation211
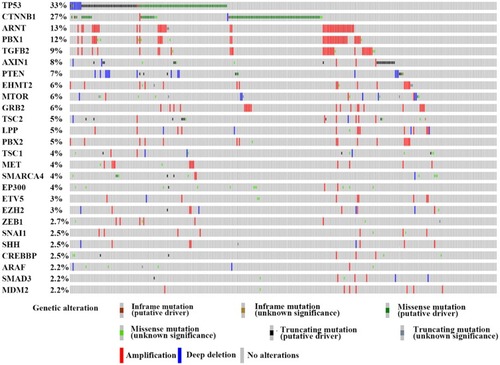
Figure 2 Genetic and epigenetic regulation mechanisms. CDH1 expression is mainly driven by Snail, zinc-finger E-box-binding (ZEB) and TWIST, which is one type of basic helix-loop-helix (bHLH) transcription factors. They bind to the E-box of CDH1 promotor to repress the expression. In this review, we focus on seven parts of regulatory pathways including mutation, CNV, DNA methylation, histone methylation, acetylation, lncRNA and miRNA. In mutation aspect, we mainly list the involvement of gene CTNNB1 and AXIN1, which both participate in the WNT/β-catenin pathway. In the WNT/β-catenin pathway, when the signaling is “on.” Axin removes from the destruction complex and β-catenin moves into the nucleus to bind to the target DNA such as SNAI1 to activate the transcription. In CNV part, we mainly list the involvement of gene ARNT, PBX1 and TGFB2. They mainly related to the WNT-β catenin and TGF-β pathway. In TGF-β pathway, TGF-β induces the SMAD pathway to activate the expression of β-catenin. In non-SMADs pathway, TGF-β regulates the Gsk-3β to change the expression of β-catenin via PI3K-Akt pathway. The HIF1ɑ factor is reported to directly regulate TWIST to have an impact on E-cadherin expression as well as Snail factor. In DNA methylation aspect, it always serves as the down-regulator of target gene expression when there exists hypermethylation area at the promotor region. Here, we list 2 genes which are reported to be regulated by DNA methylation and the recruiting factors such as Snail and Zeb1/2 as well as the recruited DNMTs. In histone methylation part, its methylation, typically at the tail of H3 and H4 histones, will epigenetically influence the gene expression due to the variant space structure. Both the H3K9me2 and H3K27me3 downregulates the expression of CDH1. In acetylation part, the balance between acetylation and deacetylation change the positive charge mainly from the lysine, thus, highly acetylated histones form more accessible chromatin and tend to be associated with active transcription. In lncRNA part, we list two main progression of its function: Decoy and Guide. LncRNAs decoy the target by binding and titrating away a protein or RNA target to negatively regulate the expression. And guide by directing the localization of specific proteins to their target to form a complex. In miRNA part, miRNAs function via base-pairing with complementary sequences (mostly in the 3ʹ-UTR) within mRNA molecules to degrade or cleave the target mRNA to silence the transcription. “ … ” represents other factors which are not listed in the figure.
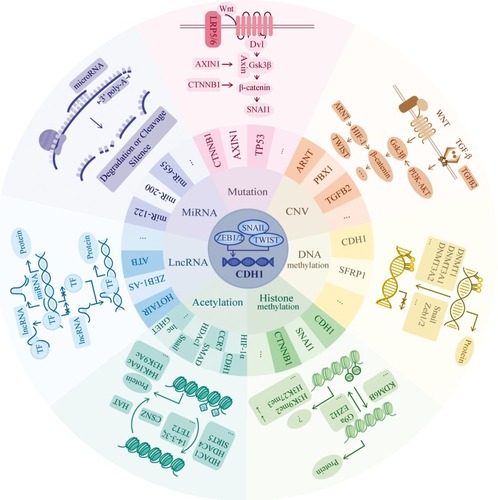
Table 1 The lncRNA And microRNA Involved In E-Cadherin Expression In HCC
