Figures & data
Table 1 Clinical Characteristics of Patients
Table 2 The Primers and Oligonucleotides Used in This Work
Figure 1 ENO1 facilitating cell proliferation was increased in oral squamous cell carcinoma (OSCC).
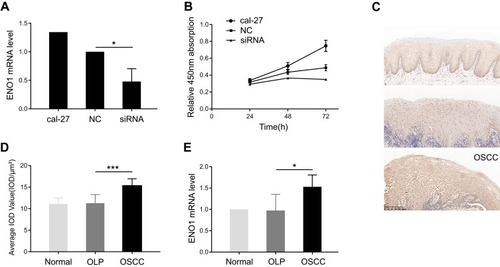
Figure 2 RNA-seq to explore the regulation network associated with ENO1.
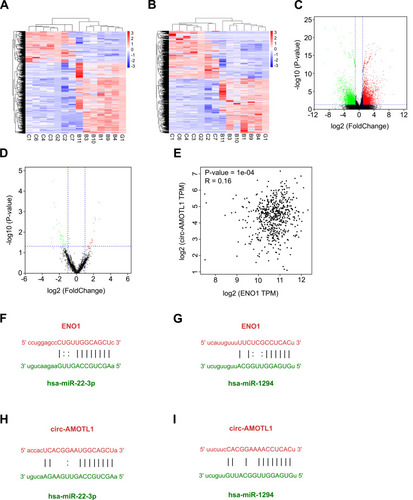
Figure 3 Circ-AMOTL1 increased in OSCC and regulated ENO1 positively.
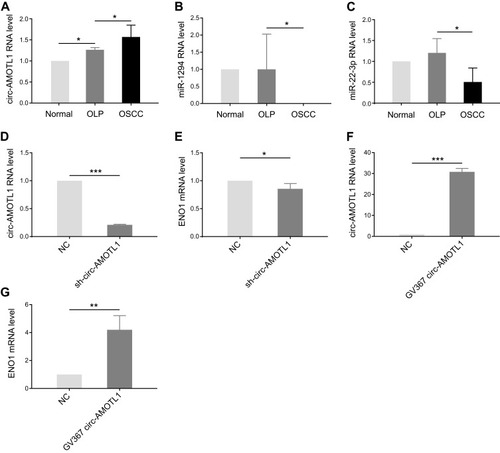
Figure 4 KEGG/GO enrichments.
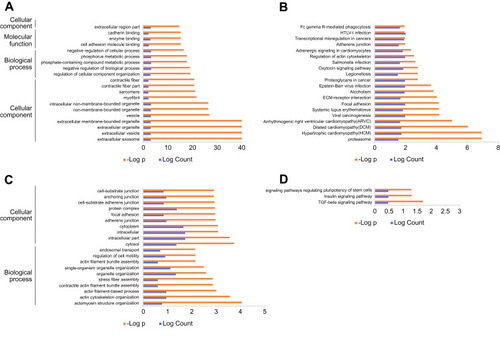
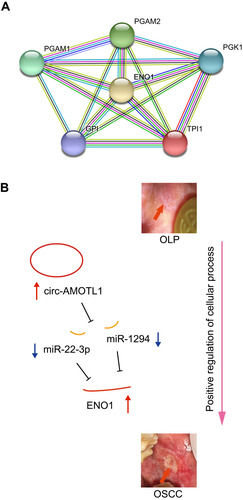
Figure 5 An ENO1 associated PPI network generated using the STRING database (A) and the hypothesis schematic diagram of the circ-AMOTL1/miR-22-3p/miR-1294/ENO1 ceRNA network during OSCC tumorigenesis (B).