Figures & data
Table 1 Characteristics of all subjects
Table 2 Primer sequences of real-time PCR
Figure 1 SR-A mRNA expression in AMs.
Abbreviations: AMs, alveolar macrophages; RT-qPCR, real-time reverse transcription quantitative polymerase chain reaction; SEM, standard error of the mean.
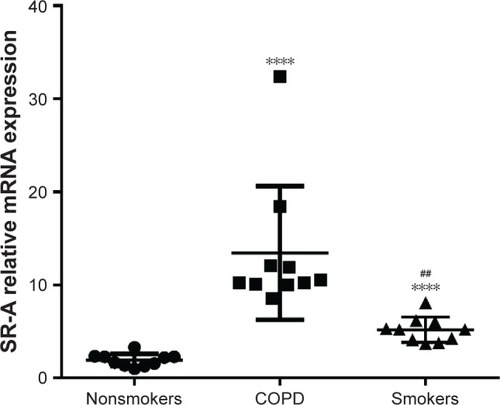
Figure 2 The distribution and expression levels of SR-A in lung tissue.
Abbreviations: SEM, standard error of the mean; SR-A, class A scavenger receptor.
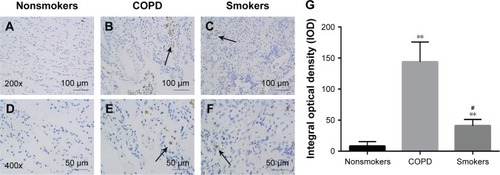
Figure 3 Confocal analyses of SR-A in AMs of COPD patients.
Abbreviation: AMs, alveolar macrophages.
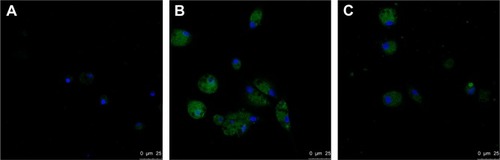
Figure 4 Cytokine secretion in BALF.
Abbreviations: BALF, bronchoalveolar lavage fluid; ELISA, enzyme-linked immunosorbent assay; SEM, standard error of the mean; TNF-α, tumor necrosis factor-alpha.
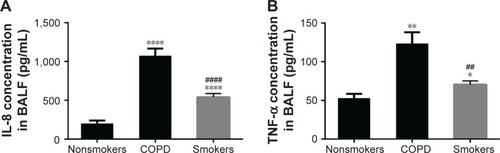
Figure 5 Relationships between SR-A mRNA, and cytokine expression levels and FEV1% predicted.
Abbreviations: AMs, alveolar macrophages; SEM, standard error of the mean; SR-A, class A scavenger receptor.
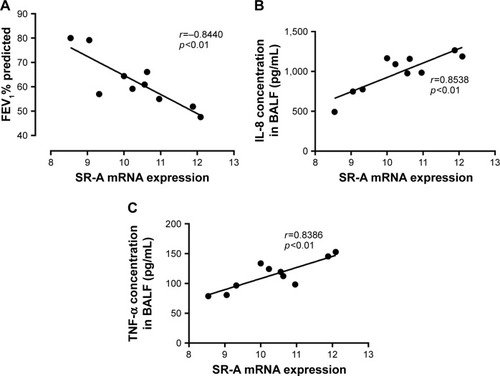
Figure 6 Western blots and RT-qPCR results of RAW-SR-A and RAW-control cells.
Abbreviations: RT-qPCR, real-time reverse transcription quantitative polymerase chain reaction.
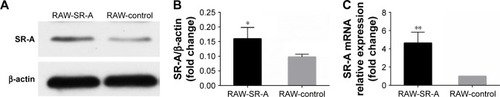
Figure 7 RT-qPCR and ELISA results of RAW-SR-A and RAW-control cells stimulated by LPS, poly(I:C), and CSE.
Abbreviations: CSE, cigarette smoke extract; ELISA, enzyme-linked immunosorbent assay; LPS, lipopolysaccharides; ns, nonsignificant; RT-qPCR, real-time reverse transcription quantitative polymerase chain reaction.
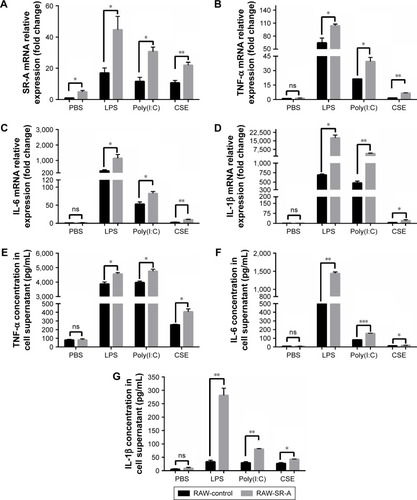
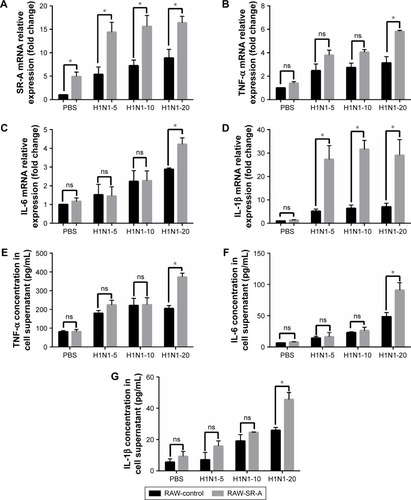
Figure 8 RT-qPCR and ELISA results of RAW-SR-A and RAW-control cells stimulated by H1N1 influenza.
Abbreviations: ELISA, enzyme-linked immunosorbent assay; ns, nonsignificant; RT-qPCR, real-time reverse transcription quantitative polymerase chain reaction.