Figures & data
Figure 1 Flowchart of study design.
Abbreviations: DE, differentially expressed; FC, fold change; RPM, reads per million; FPKM, fragments per kilobase of transcript per million mapped reads; KEGG, Kyoto Encyclopedia of Genes and Genomes.
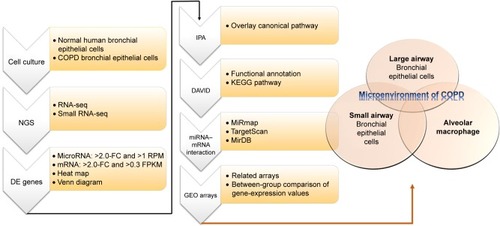
Table 1 Dysregulated genes with potential microRNA–mRNA interactions in COPD bronchial epithelial cells
Figure 2 Identification of genes with potential microRNA–mRNA interactions in COPD bronchial epithelial cells.
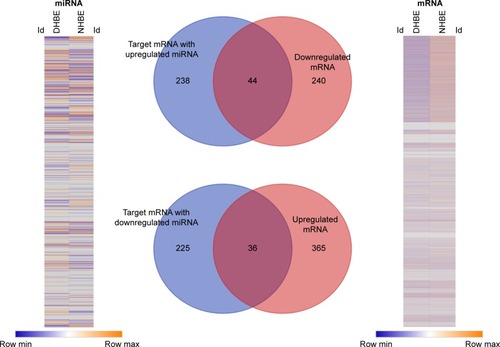
Table 2 Downregulated genes with microRNA–mRNA interactions and their functions
Figure 3 Functional analysis of dysregulated genes identified in COPD epithelial cells by Ingenuity Pathway Analysis (IPA).
Notes: The 80 identified dysregulated genes with potential microRNA–mRNA interactions were analyzed by IPA. (A) Pathways related to 44 downregulated genes; (B) pathways related to 36 upregulated genes.
Abbreviations: TR, thyroid hormone receptor; RXR, retinoid X receptor; PTEN, phosphatase and tensin homolog; FAK, focal adhesion kinase; NFAT, nuclear factor of activated T-cell.
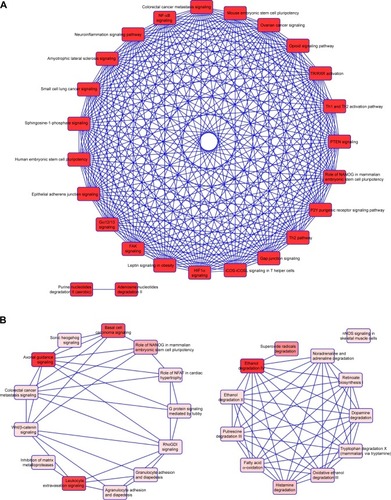
Figure 4 Possible mechanisms of dysregulated genes identified in COPD epithelial cells analyzed by Database for Annotation, Visualization, and Integrated Discovery (DAVID).
Notes: Functional annotation of the 44 downregulated genes was determined by gene ontology using DAVID. These 44 genes are involved in the functioning of membrane, transmembrane helix, transmembrane, nucleotide bonding, cell adhesion, calcium, disulfide bonds, and signals.
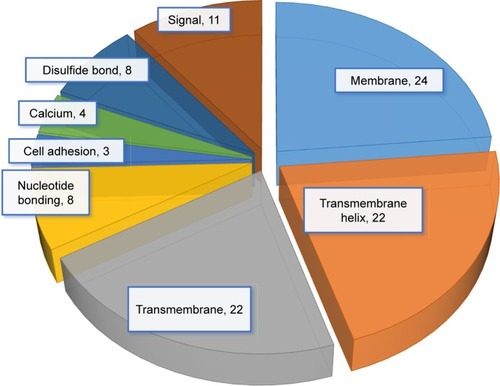
Figure 5 Gene Expression Omnibus (GEO) database analysis of 18 downregulated genes with potential microRNA–mRNA interactions in COPD small airway.
Abbreviation: NS, not significant.
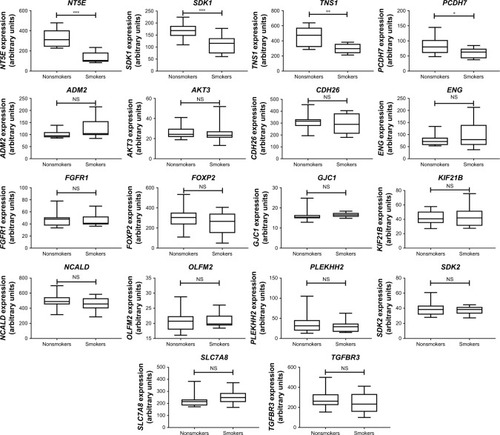
Figure 6 Gene Expression Omnibus database analysis of the four genes downregulated in COPD small airway, large airway, and alveolar macrophages.
Abbreviation: NS, not significant.
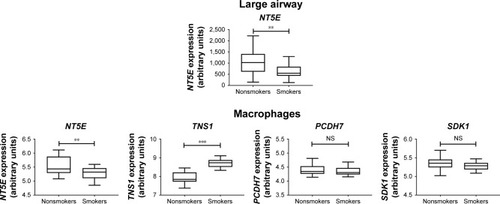
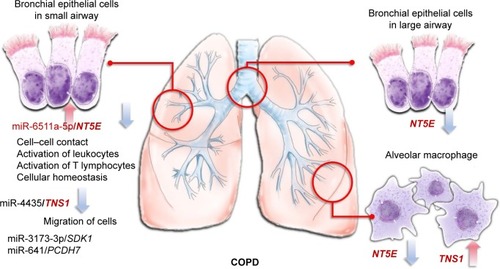
Figure 7 MicroRNA–mRNA interactions in the microenvironment of COPD.