Figures & data
Table 1 Five BLAST aligned templates of HSPB8 with identity, query coverage, and E-values
Figure 1 Root mean square deviation (RMSD) versus time.
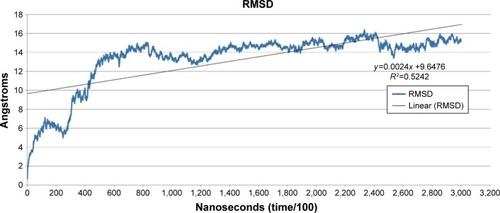
Figure 2 Root mean square fluctuation (RMSF), showing residue fluctuations.
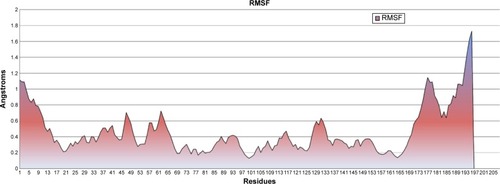
Figure 3 Average root mean square deviation (RMSD) values per frame.
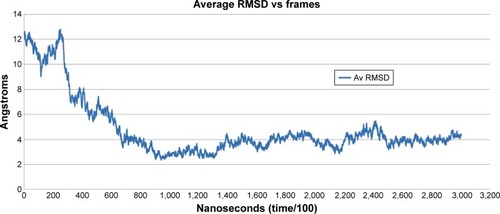
Table 2 Compounds investigated in this study
Figure 5 2-D structures of investigated drugs.
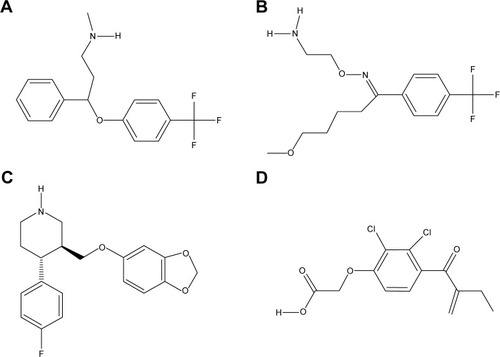
Table 3 Bioinformative details and ligand properties of top four screened compounds
Figure 6 2-D structures of novel molecules.
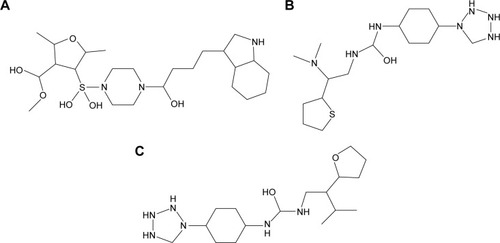
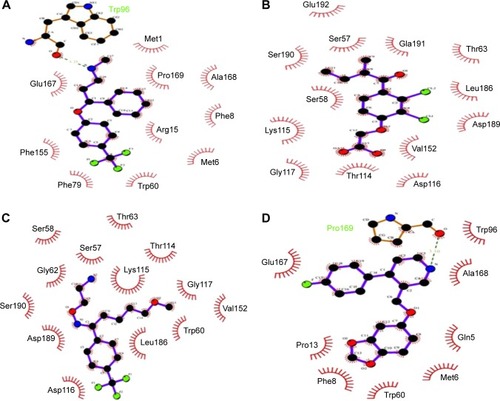
Figure 7 The potential binding interactions of investigated drugs through LigPlot.