Figures & data
Figure 1 Strategy of dual-target inhibitor discovery against MyD88 and Nur77.
Abbreviations: TCM, traditional Chinese medicine; CP1, Compound 1; CP2, Compound 2.
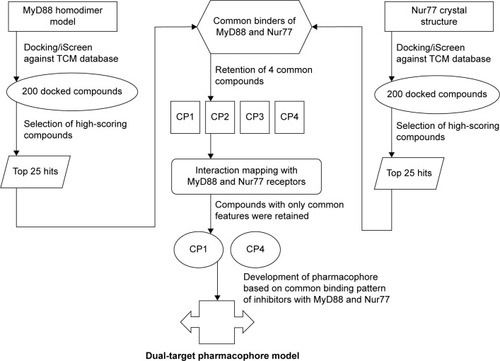
Figure 2 Superimposed structures of the docked MyD88 dimers showing complete overlap of the docked complexes.
Abbreviation: GRAMM-X, Global Range Molecular Matching.
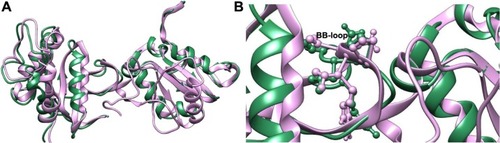
Table 1 The top 25 virtual screening hits and their scores for MyD88 from TCM database integrated iScreen program
Table 2 The top 25 virtual screening hits and their scores for Nur77 from TCM Database integrated iScreen program
Table 3 Four compounds selected on the basis of common binding to both MyD88 and Nur77 targets
Figure 3 Mode of inhibitor docking in the MyD88 binding site.
Abbreviations: MyD88, myeloid differentiation primary response protein 88; CP1, Compound 1; CP2, Compound 2; CP3, Compound 3; CP4, Compound 4.
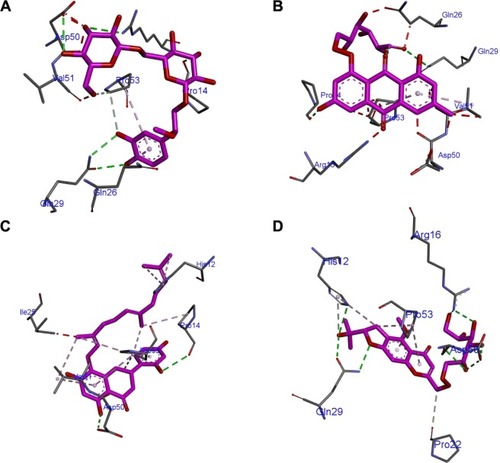
Table 4 Corresponding sequence numbers of residues in Uniprot database as well as in the docked poses for Myd88 and Nur77
Figure 4 Mode of inhibitor docking in the Nur77 binding site.
Abbreviations: CP1, Compound 1; CP2, Compound 2; CP3, Compound 3; CP4, Compound 4.
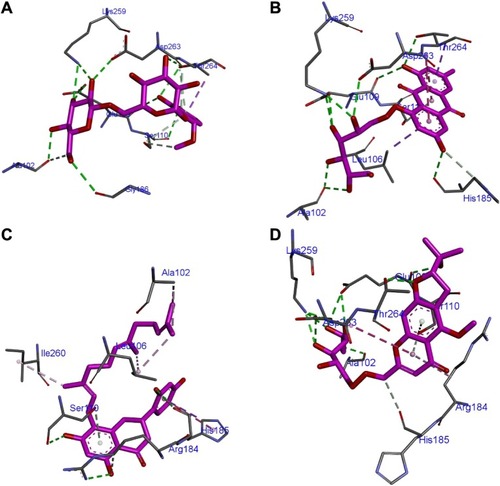
Figure 5 Two-dimensional representation of inhibitor binding in the MyD88 binding site.
Abbreviations: CP1, Compound 1; CP2, Compound 2; CP3, Compound 3; CP4, Compound 4.
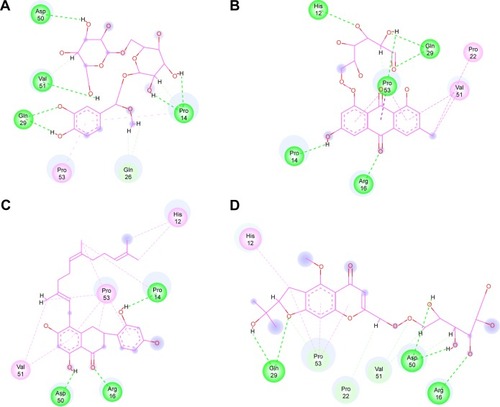
Figure 6 Two-dimensional representation of inhibitor binding in the Nur77 binding site.
Abbreviations: CP1, Compound 1; CP2, Compound 2; CP3, Compound 3; CP4, Compound 4.
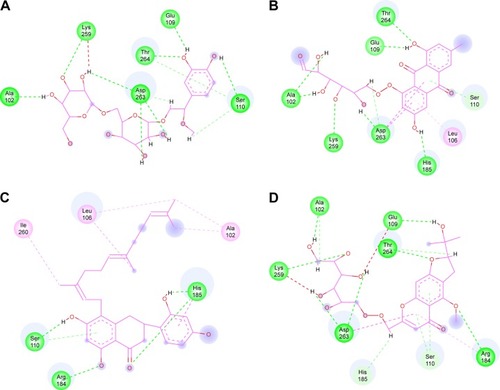
Figure 7 Pharmacophore mapping of the docked complexes.
Abbreviations: CP1, Compound 1; CP4, Compound 4.
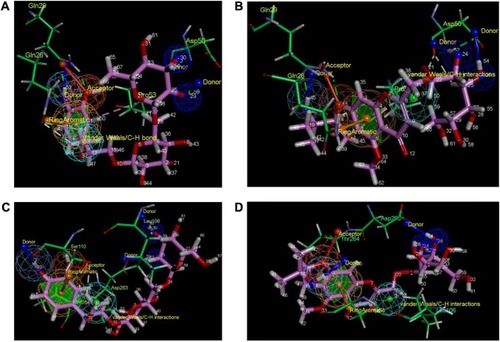
Table 5 The similarity between CP1 and CP4 for binding same residues in MyD88 and Nur77 targets, respectively, via a similar set of atoms
Figure 8 The final pharmacophore model based on MyD88 and Nur77.
Abbreviations: CP1, Compound 1; CP4, Compound 4.
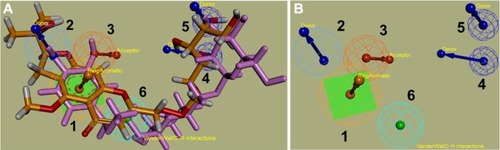
Table 6 Distances between various pharmacophore points in the final pharmacophore model
