Figures & data
Figure 1 Transcriptomics and genetic alteration analyses of cytoskeleton regulators.
Abbreviations: TCGA, The Cancer Genome Atlas; IP, interacting proteins; OR, organizing and biogenesis; P-D, polymerization or depolymerization; CC, cortical cytoskeleton; SP, spindle organization and biogenesis; AX, axon and dendrites formation; FL, filopodia; G, growth cones; LAM, lamellipodia; MV, microvilli; P, pseudopodia; RF, ruffles; CS-S-P, cell shape–size–polarity; CMM, cell motility or migration; M, mitosis; CK, cytokinesis; CA, cytoskeleton adaptors; Cal, calmodulins and calcineurins; G-Prot, G-protein signaling; Kin&Pho, kinases and phosphatases.
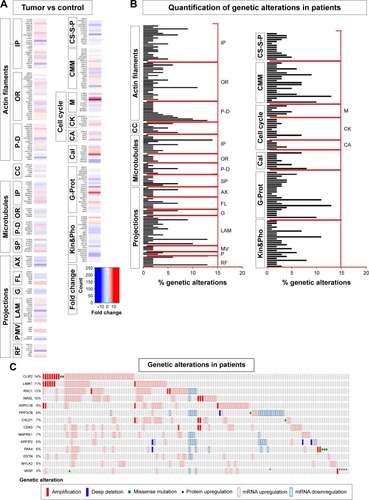
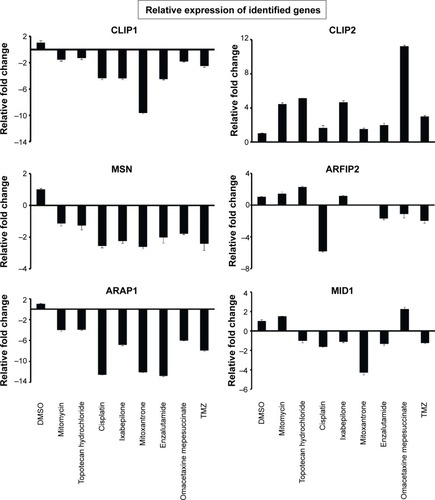
Figure 2 Transcriptomics data shown as a color code representing fold change in expression of 85 genes grouped into ten groups with abbreviated subgroups, in GBM samples compared to neural stem cells (NSCs).
Abbreviations: GBM, glioblastoma multiforme; IP, interacting proteins; OR, organizing and biogenesis; P-D, polymerization or depolymerization; CC, cortical cytoskeleton; SP, spindle organization and biogenesis; AX, axon and dendrites formation; FL, filopodia; G, growth cones; LAM, lamellipodia; MV, microvilli; P, pseudopodia; RF, ruffles; CS-S-P, cell shape–size–polarity; CMM, cell motility or migration; M, mitosis; CK, cytokinesis; CA, cytoskeleton adaptors; Cal, calmodulins and calcineurins; G-Prot, G-protein signaling; Kin&Pho, kinases and phosphatases.
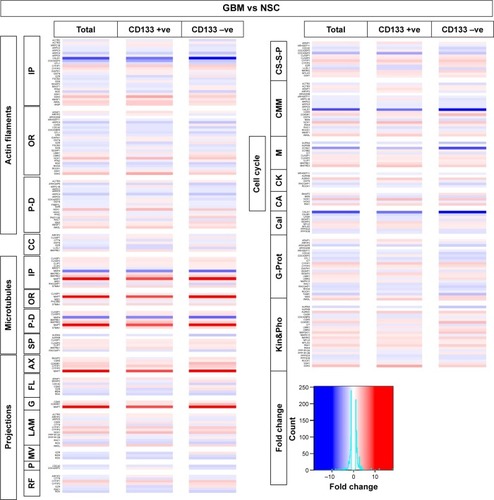
Figure 3 Transcriptomics and patient survival plots of cytoskeletal regulators.
Abbreviations: GBM, glioblastoma multiforme; IP, interacting proteins; OR, organizing and biogenesis; P-D, polymerization or depolymerization; CC, cortical cytoskeleton; SP, spindle organization and biogenesis; AX, axon and dendrites formation; FL, filopodia; G, growth cones; LAM, lamellipodia; MV, microvilli; P, pseudopodia; RF, ruffles; CS-S-P, cell shape–size–polarity; CMM, cell motility or migration; M, mitosis; CK, cytokinesis; CA, cytoskeleton adaptors; Cal, calmodulins and calcineurins; G-Prot, G-protein signaling; Kin&Pho, kinases and phosphatases.
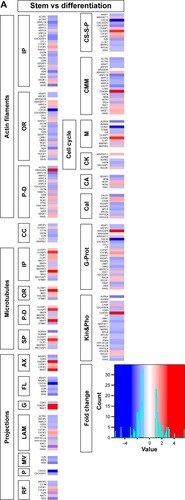
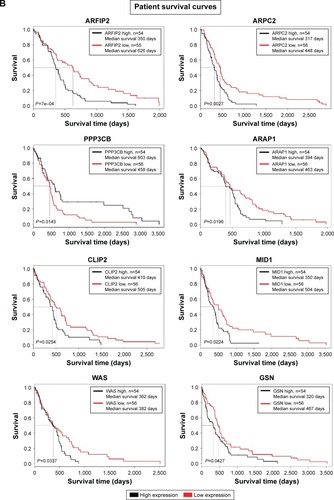
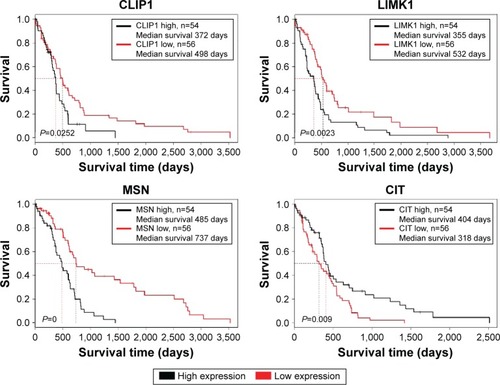
Figure 4 Cell viability screen using FDA approved oncology drugs.
Abbreviation: DMSO, dimethyl sulfoxide.
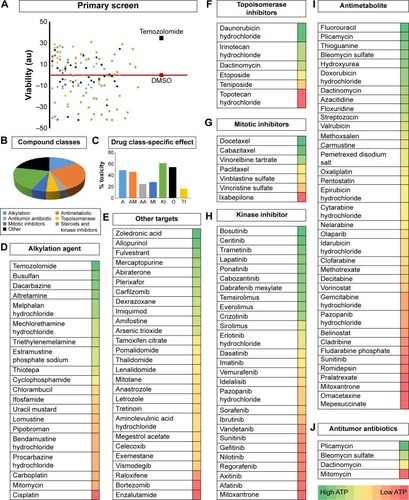
Figure 5 Cell viability analyses of temozolomide and other compounds.
Abbreviations: IC50, half maximal inhibitory concentration; GBM, glioblastoma multiforme; DMSO, dimethyl sulfoxide.
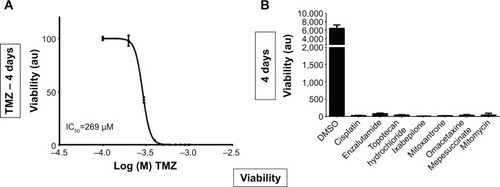
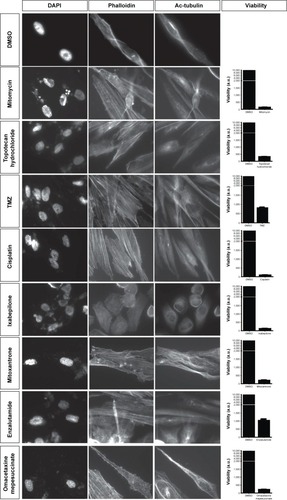
Figure 6 Staining of GBM cells with DAPI, phalloidin, and antiacetylated tubulin antibody after 2 days treatment with DMSO or different compounds.
Abbreviations: GBM, glioblastoma multiforme; DAPI, 4′,6-diamidino-2-phenylindole; DMSO, dimethyl sulfoxide; Ac, antiacetylated.