Figures & data
Figure 1 Mechanism of MG detoxification utilizing glyoxalase system enzymes.
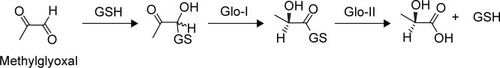
Figure 2 Takasawa perception of the expected pharmacophore and SAR of flavonoids as Glo-I inhibitors.
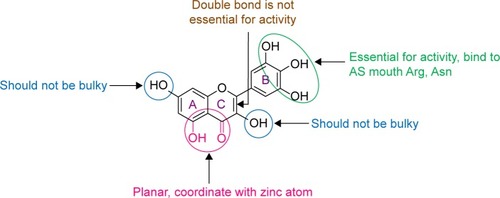
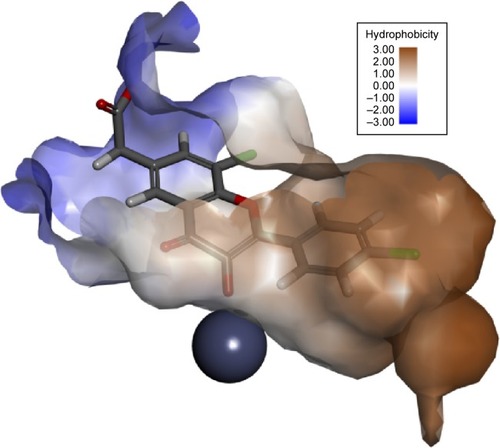
Figure 3 Active site of the Glo-I enzyme.
Abbreviation: Glo-I, glyoxalase-1.
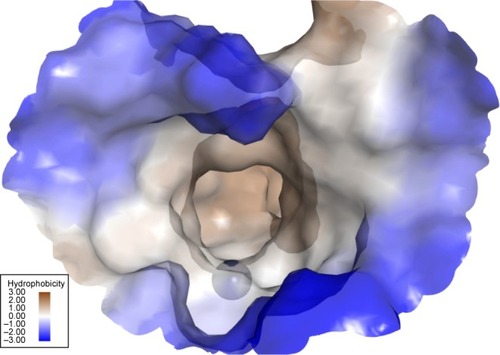
Figure 4 Query used for extracting compounds from the AldrichCPR database using the Ligand Pharmacophore Mapping protocol.
Table 1 Chemical structures of retrieved hits from AldrichCPR, their in vitro enzyme assay results, and corresponding in silico docking scores