Figures & data
Figure 1 The identification of the triazole compound that activated autophagy.
Abbreviations: ΔC-TBP, C-terminus-truncated TBP; DMSO, dimethyl sulfoxide; EGFP, enhanced green fluorescence protein; h, hours.
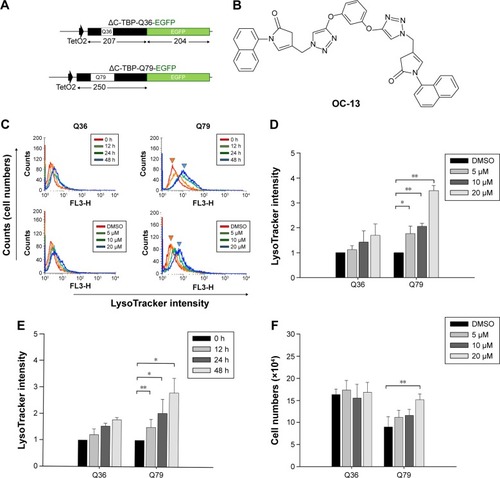
Figure 2 OC-13 activated autophagic flux and dissolved the insoluble Q79-EGFP aggregates.
Abbreviations: D, DMSO; DMSO, dimethyl sulfoxide; EGFP, enhanced green fluorescence protein; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; h, hours.
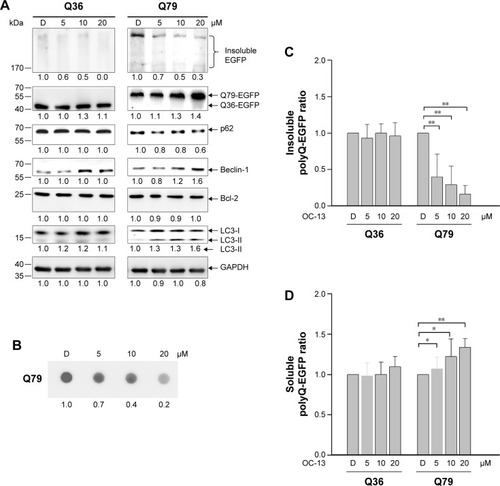
Figure 3 Nucleus exclusion and autophagic amelioration of Q79-EGFP aggregates.
Abbreviations: D, DMSO; DAPI, 4′,6-diamidino-2-phenylindole; DMSO, dimethyl sulfoxide; EGFP, enhanced green fluorescence protein; h, hours.
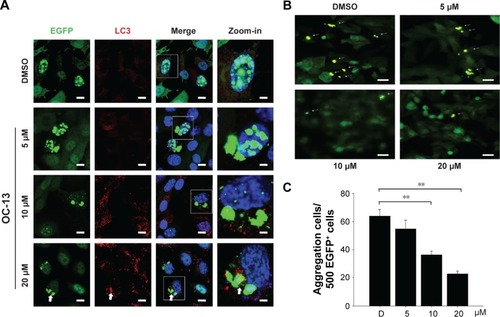
Figure 4 The autophagy pathway is related to JNK signaling activation.
Abbreviations: Baf A1, bafilomycin A1; D, DMSO; DMSO, dimethyl sulfoxide; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; JNK1/2, c-Jun N-terminal protein kinase 1 and 2; h, hour.
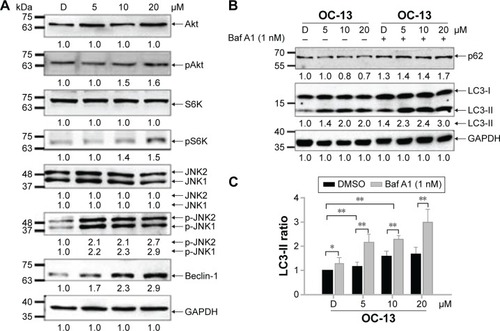
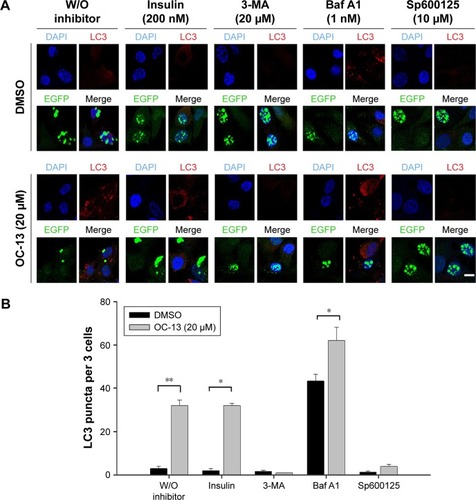
Figure 5 Inhibition autophagy flux blocked clearance of the Q79-EGFP aggregates.
Abbreviations: Baf A1, bafilomycin A1; D, DMSO; DMSO, dimethyl sulfoxide; EGFP, enhanced green fluorescence protein; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; JNK, c-Jun N-terminal protein kinase; 3-MA, 3-methyladenine; h, hour; W/O, without.
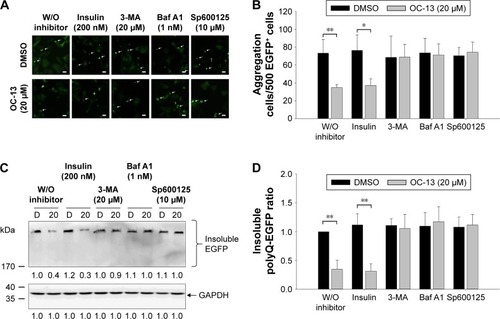
Figure 6 Inhibition lysosome fusion with autophagosome impaired clearance of the Q79-EGFP aggregates.
Abbreviations: Baf A1, bafilomycin A1; DAPI, 4′,6-diamidino-2-phenylindole; DMSO, dimethyl sulfoxide; EGFP, enhanced green fluorescence protein; 3-MA, 3-methyladenine; h, hour; W/O, without.