Figures & data
Table 1 Protein sequences used in the homology modeling of the NS5 RdRp for all four dengue virus serotypes
Figure 1 Multiple sequence alignment of NS5 RNA-dependent RNA polymerases of the different dengue virus serotypes (above) and the corresponding percent identitity matrix (below).
Notes: Pink boxes indicate the aminoacid position of M343, T413, and R737 for DENV-3 considering the full length of NS5 (MTase N-terminal + RdRp C-terminal) or the equivalent position for the other serotypes. Yellow boxes indicate missing residues in the crystallographic data. Orange boxes indicate the GDD motif. For each sequence, the UniProt code for the full polyprotein of the virus | SEROTYPE is indicated.
Abbreviation: NS5, nonstructural protein 5.

Table 2 Molecular docking analysis for NITD-related and potential inhibitor compounds (candidate molecules in this study) at the binding site of NS5 RdRp located in the RNA template tunnel
Table 3 Molecular docking analysis for natural compounds
Table 4 Physicochemical parameters for selected compounds based on molecular docking analysis
Figure 2 Comparison of the free energy variation (ΔG) for selected compounds based on molecular docking analysis against NS5 RdRps of all four serotypes of dengue virus (A). ΔG for NITD compounds is also included. (B) The molecular structures of the compounds represented in (A); the PubChem ID or the SuperNatural 2 ID is close to the structure of each compound.
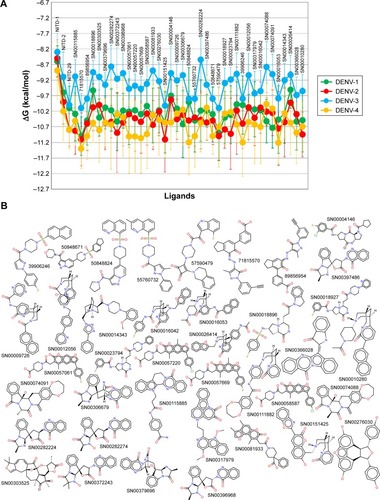
Figure 3 3D structure of the proteins NS5 RNA-dependent RNA polymerases of DENV-4 (A and B) and DENV-2 (C and D) showing the binding sites (left), the binding cavity (middle), and the main residues involved in the ligand–protein interaction of compound 50848824 (A), 71815570 (B), SN00151425 (C), and SN00010280 (D). Structure visualization was by PyMol 1.8.2.0.
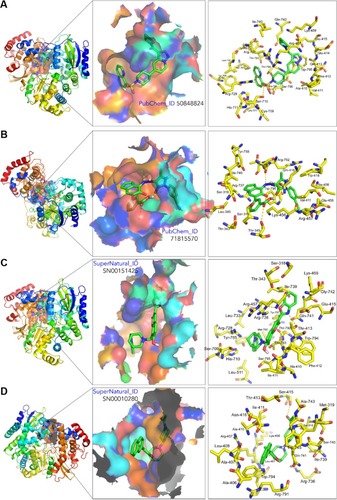
Table 5 Predicted molecular pharmacokinetic properties of selected compounds against the NS5 RdRp
Table 6 Predicted toxicity assessment of selected compounds against the NS5 RdRp
