Figures & data
Figure 1 Design of compounds to bind to hIL-6R.
Abbreviations: 3D-ACD, three-dimensional-Available Chemicals Directory; MDDR, MDL Drug Data Report.
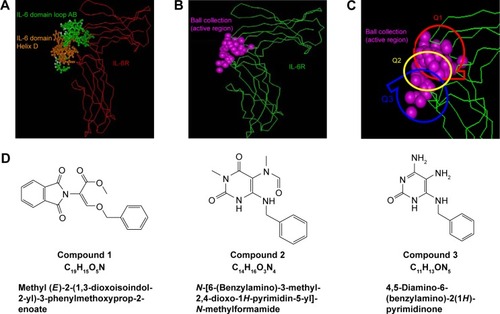
Table 1 Key residues and binding energy between hIL-6R and compounds
Figure 2 Theoretical analysis of candidate compounds binding to hIL-6R.
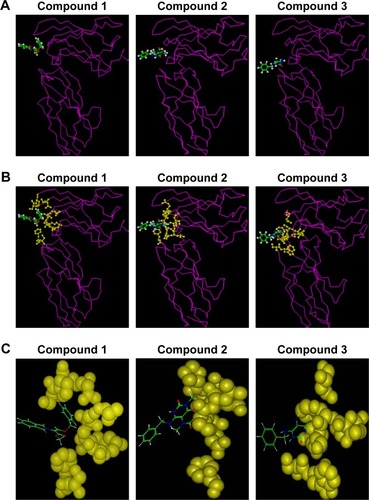
Figure 3 Competing ELISA analysis of compounds at different concentrations to bind to hIL-6R.
Abbreviations: ELISA, enzyme-linked immunosorbent assay; NEG, negative; PBS, phosphate-buffered saline; DMSO, dimethyl sulfoxide; SD, standard deviation; OD, optical density.
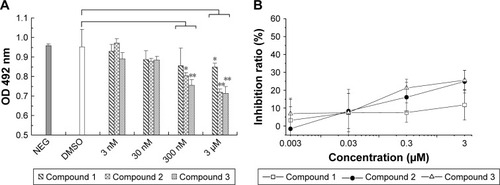
Figure 4 The inhibition effect of compounds on XG-7 cells by cell proliferation assays.
Abbreviations: MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; NEG, negative; DMSO, dimethyl sulfoxide; POS, positive; SD, standard deviation; OD, optical density; 3H-Tdr, 3H tritiated thymidine; CPM, counts per minute; CPS, counts per second.
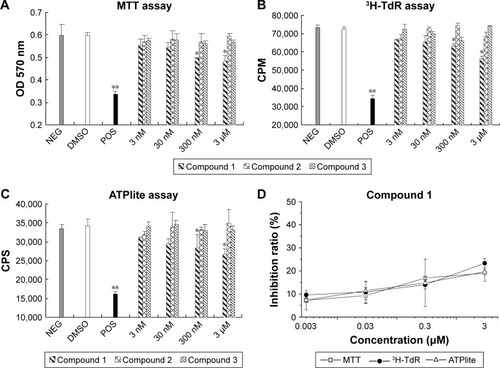
Figure 5 Compound 1 induced XG-7 cell apoptosis.
Abbreviations: NEG, negative; DMSO, dimethyl sulfoxide; POS, positive.
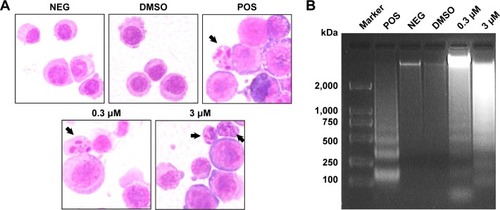
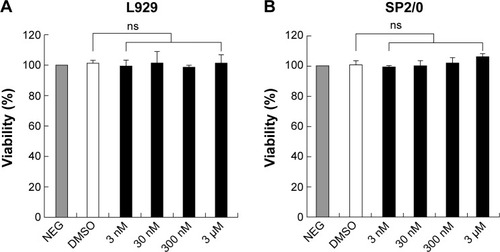
Figure 6 The detection of cytotoxic effect of compound 1 with MTT assay.
Abbreviations: MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; SD, standard deviation; NEG, negative; DMSO, dimethyl sulfoxide; ns, not significant.