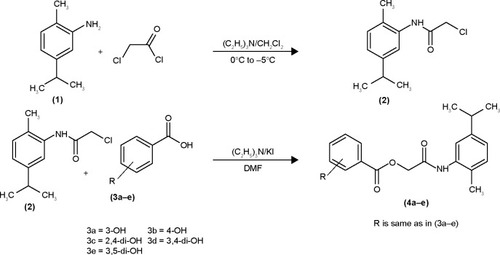
Figures & data
Table 1 Inhibitory effects of amide derivatives 4a–e and 6a–e on mushroom tyrosinase and porcine pancreas elastase
Table 2 Inhibitory effects of amide derivatives 4a–e and 6a–e on human tyrosinase (from melanoma cells) free radical scavenging
Table 3 Kinetic parameters of mushroom tyrosinase for L-DOPA activity in the presence of different concentration of compounds 4c, 6a, 6b and 6d
Figure 1 Lineweaver–Burk plots for inhibition of tyrosinase in the presence of amide 4c.
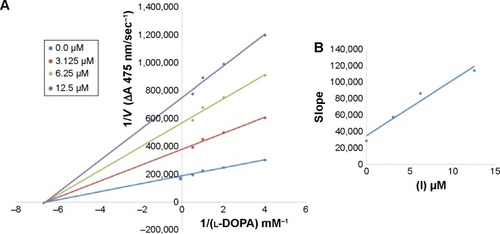
Figure 2 Lineweaver–Burk plots for inhibition of tyrosinase in the presence of amide 6a.
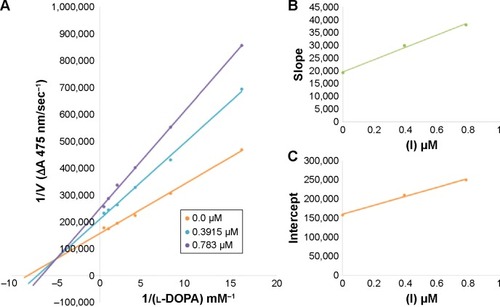
Figure 3 Lineweaver–Burk plots for inhibition of tyrosinase in the presence of amide 6b.
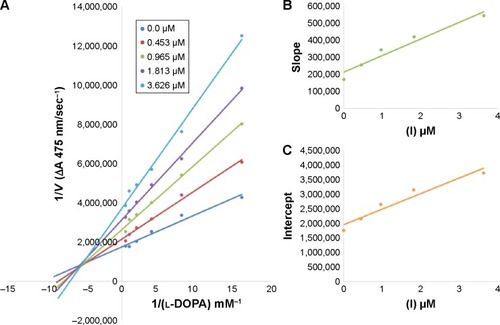
Figure 4 Lineweaver–Burk plots for inhibition of tyrosinase in the presence of amide 6d.
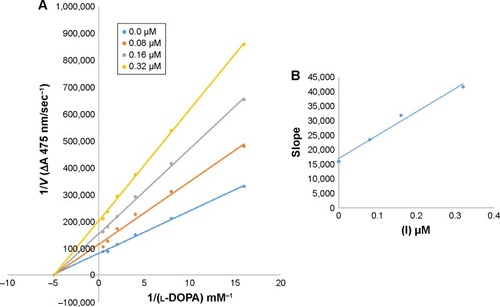
Figure 5 Effect of various doses of mushroom tyrosinase on its activity for the catalysis of l-DOPA against different concentration of inhibitor 6d.
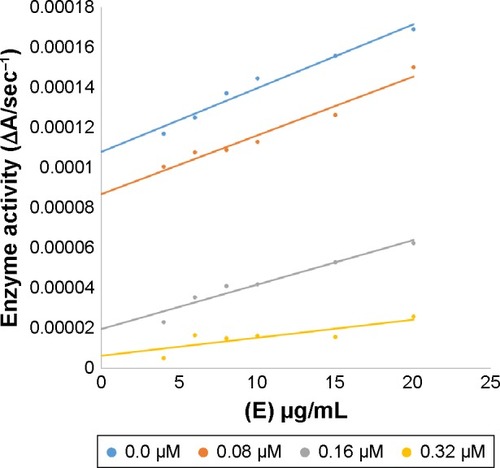
Figure 6 Effect of inhibitor 6d on pigmentation of zebrafish.
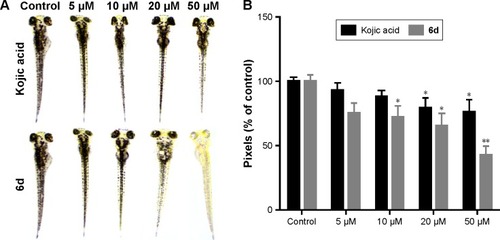
Figure 7 Inhibitory effects of 6d and kojic acid on melanin contents.
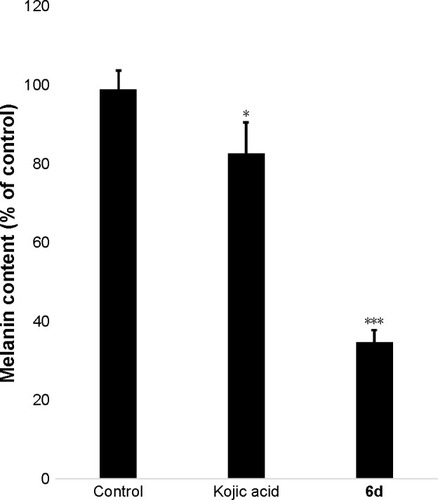
Figure 8 Zebrafish embryos (48 hpf) were treated with 10, 20 and 50 µM of 6d.
Abbreviation: hpf, hours post-fertilization.
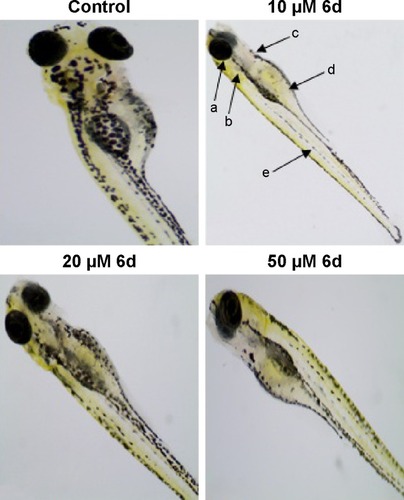
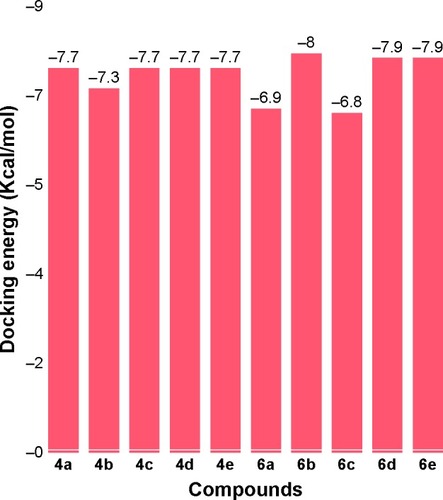
Table 4 Chemo-informatics evaluation of the synthesized compounds
Figure 10 Docking interactions between 6d and target protein.
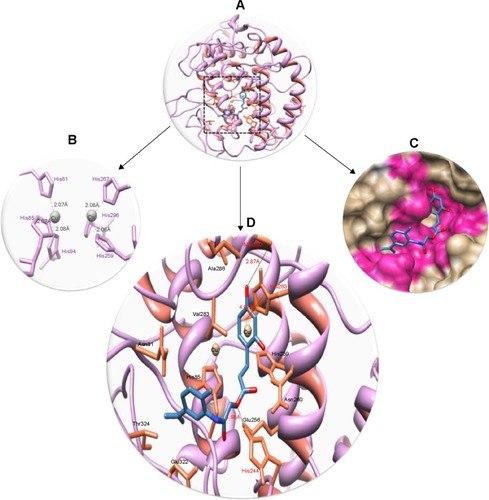
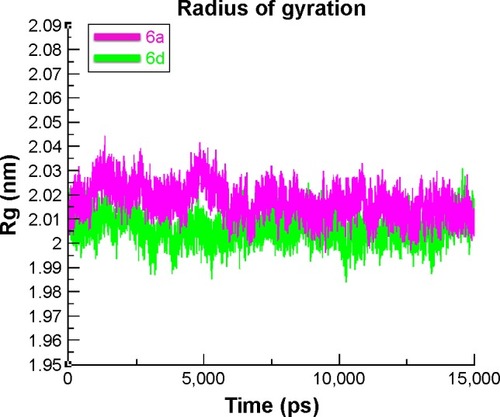
Figure 11 RMSD graph of 6a and 6d at 15 ns.
Abbreviation: RMSD, root mean square deviation.
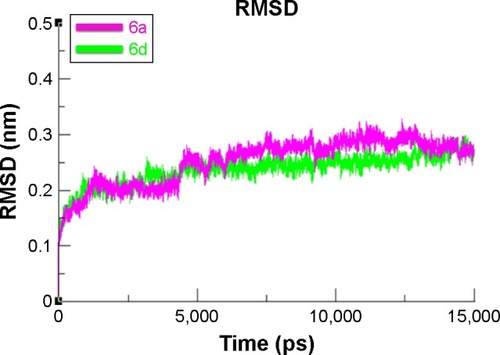
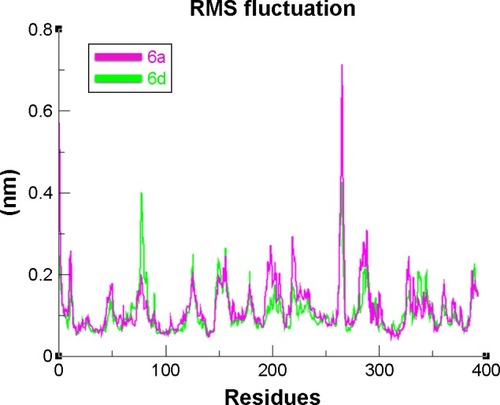
Figure 12 RMSF graph of 6a and 6d at 15 ns.
Abbreviation: RMSF, root mean square fluctuation.