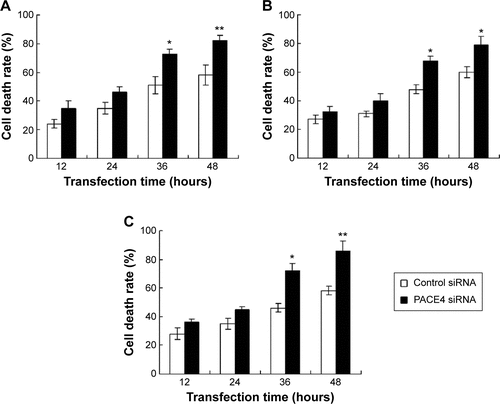
Figures & data
Figure 1 Proliferation of prostate cancer cells were inhibited by PACE4 siRNA.
Abbreviations: CCK, cell-counting kit; OD, optical density; SD, standard deviation; siRNA, small interfering RNA.
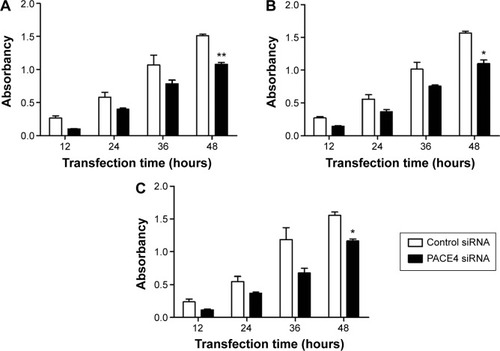
Figure 2 PACE4 siRNA induces cell cycle arrest in prostate cancer cells.
Abbreviation: siRNA, small interfering RNA.
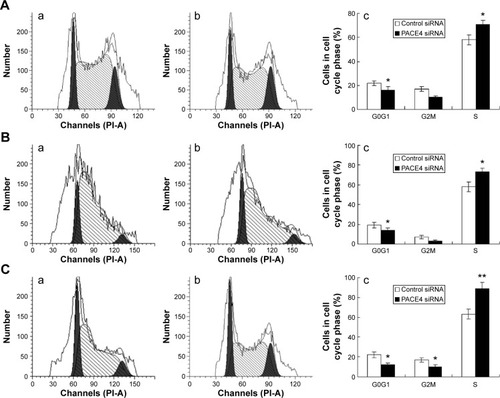
Figure 3 PACE4 siRNA significantly increased apoptosis of prostate cancer cells.
Abbreviation: siRNA, small interfering RNA.
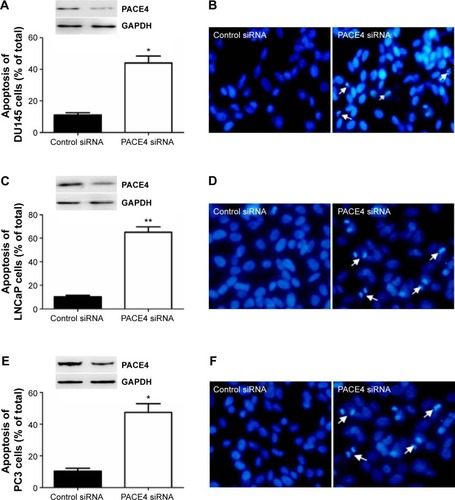
Figure 4 Western blot analysis of apoptotic-related proteins after PACE4 siRNA transfection.
Abbreviations: SD, standard deviation; siRNA, small interfering RNA.
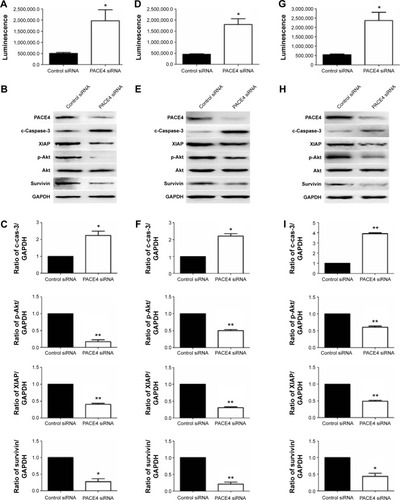
Figure 5 Regulation of mediators in the mitochondrial pathway in apoptotic cells by PACE4 siRNA.
Abbreviations: Cyto c, cytochrome c; SD, standard deviation; siRNA, small interfering RNA; vs, versus.
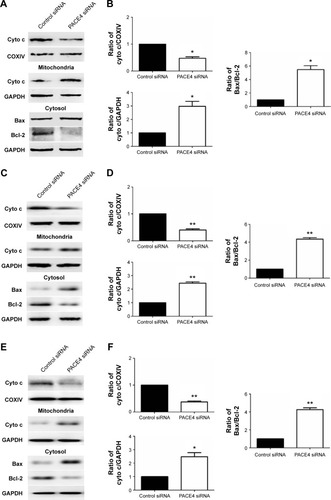
Figure 6 Effects of PACE4 siRNA on ER stress-associated proteins in prostate cancer cells.
Abbreviations: ER, endoplasmic reticulum; SD, standard deviation; siRNA, small interfering RNA.
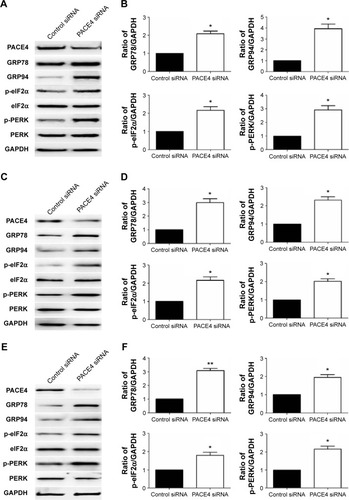
Figure 7 Observation of apoptosis in PACE4 gene knockdown prostate cancer cell lines.
Abbreviations: ER, endoplasmic reticulum; SD, standard deviation.
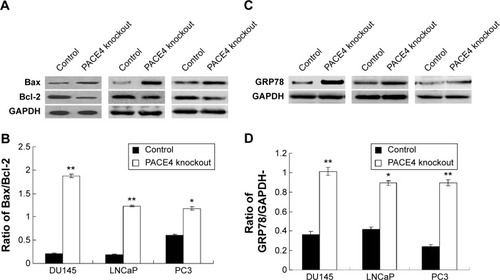
Figure S1 Observation of the cell death, using MMT assay.
Abbreviations: MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; OD, optical density; SD, standard deviation; siRNA, small interfering RNA.