Figures & data
Table 1 Primers used in the construction of IFN-λ analogs and qPCR assays
Figure 1 Design of IFN-λ analogs.
Abbreviation: IFN, interferon.
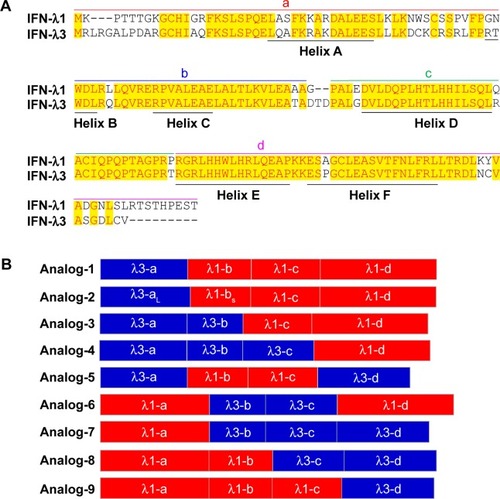
Figure 2 Analysis and characterization of expressed proteins.
Abbreviations: IFN, interferon; IPTG, isopropyl-β-D-thiogalactopyranoside; PEG-IFN, pegylated-interferon; SDS-PAGE, polyacrylamide gel electrophoresis.
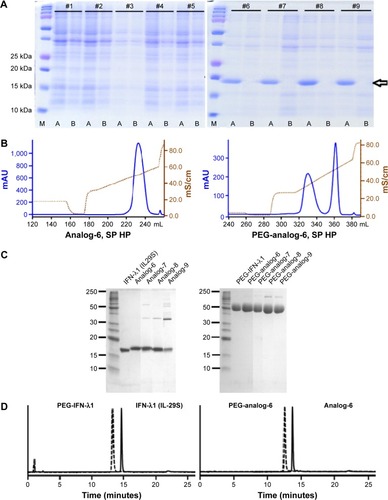
Table 2 Characterization of purified proteins by LC/Q-TOF/MS
Figure 3 Stimulation of antiviral gene expression.
Abbreviations: IFN, interferon; PEG-IFN, pegylated-interferon; qRT-PCR, quantitative real-time polymerase chain reaction.
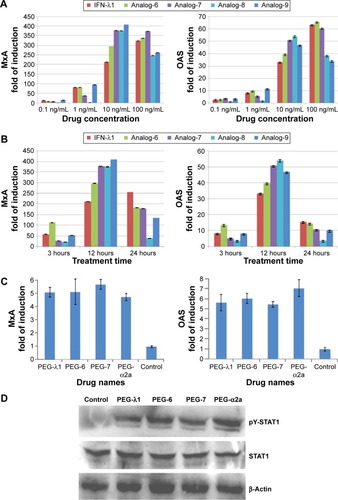
Figure 4 Activation of the ISRE-luciferase reporter.
Abbreviations: IFN, interferon; ISRE, IFN-stimulated response element; PEG-IFN, pegylated-interferon.
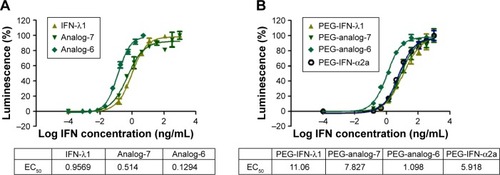
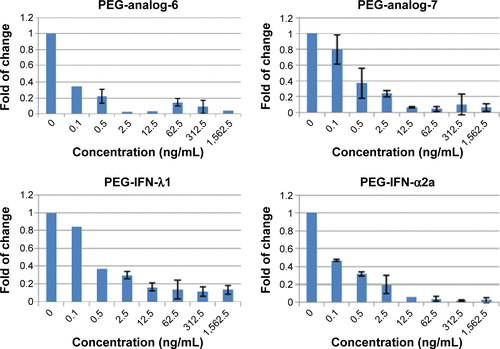
Figure 5 Anti-HCV assay.
Abbreviations: HCV, hepatitis C virus; IFN, interferon; PEG-IFN, pegylated-interferon.
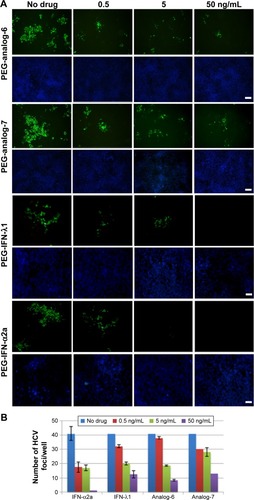
Figure 6 Quantification of anti-HCV and anti-H3N2 potency.
Abbreviations: HCV, hepatitis C virus; IFN, interferon; ELISA, enzyme-linked immunosorbent assay; IC50, half maximal inhibitory concentration; PEG-IFN, pegylated-interferon.
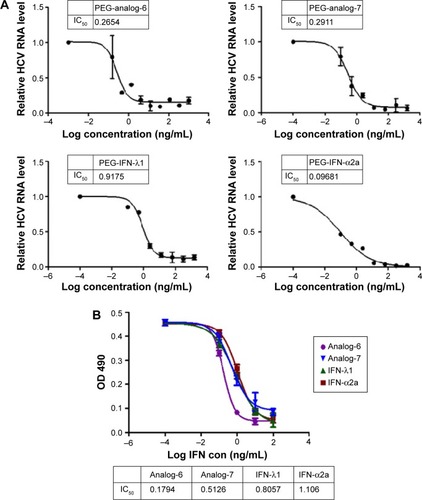
Figure S1 Amino acid sequences of the nine designed analogs.

Figure S2 Stability of analog protein preparations.
Abbreviation: HPLC, high-performance liquid chromatography.
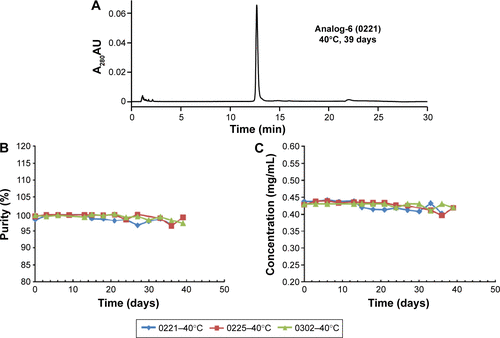
Figure S3 qRT-PCR analysis of MxA and OAS gene expression.
Abbreviations: IL, interleukin; qRT-PCR, quantitative real-time polymerase chain reaction.
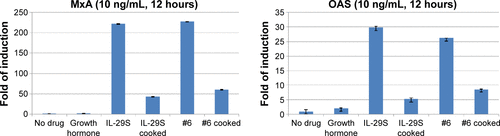
Figure S4 MTT assay.
Abbreviations: IFN, interferon; ISRE, IFN-stimulated response element; PEG-IFN, pegylated-interferon.
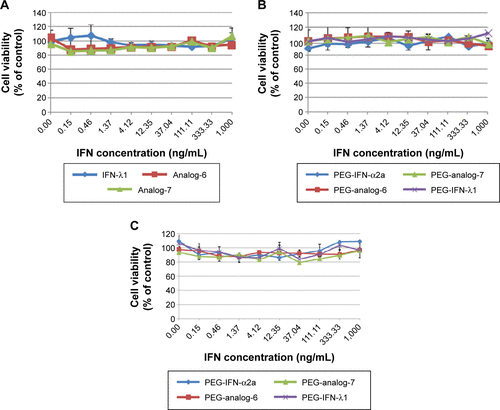
Figure S5 qRT-PCR measurement of changes in HCV RNA levels.
Abbreviations: HCV, hepatitis C virus; IFN, interferon; qRT-PCR, quantitative real-time polymerase chain reaction; PEG-IFN, pegylated-interferon.