Figures & data
Figure 2 Characterization of isolated compound of Ocimum sanctum (quercetol) by (A) UV spectra, (B) FTIR spectra, (C) (a) NMR spectra, (b) structure of quercetol, and (D) mass spectra.
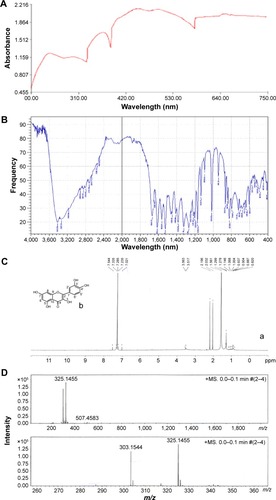
Figure 3 Morphology of HepG2 cells.
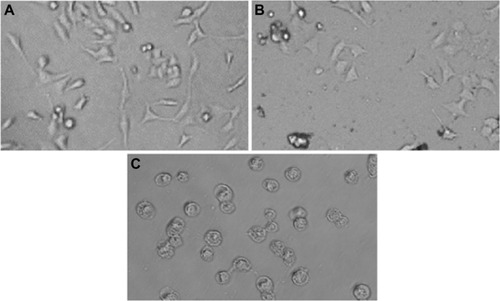
Figure 4 Toxic effect of quercetol in HepG2 cells as determined by (A) MTT and (B) NRU tests.
Abbreviations: ANOVA, analysis of variance; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; NRU, neutral red uptake; SE, standard error.
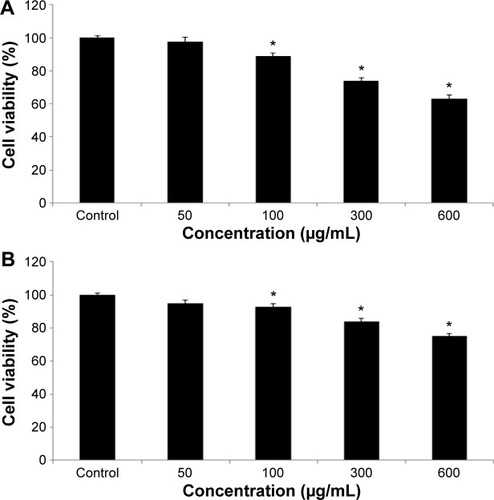
Figure 5 Impact of quercetol on the MMP of HepG2 cells.
Abbreviations: ANOVA, analysis of variance; MPP, mitochondrial membrane potential; SE, standard error.
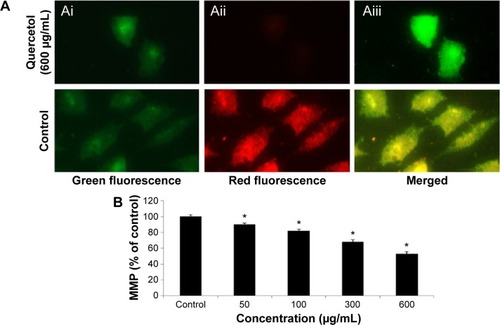
Figure 6 Induction of condensed chromosome and caspase-3 enzyme activity in HepG2 cells after exposure to quercetol.
Abbreviations: ANOVA, analysis of variance; SE, standard error.
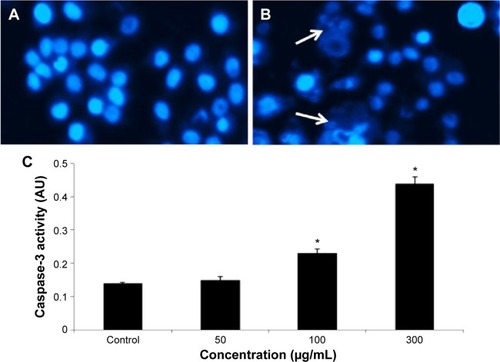
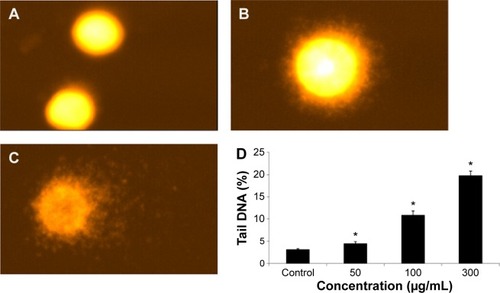
Figure 7 Fragmentation of DNA in HepG2 cells due to 24 hour quercetol exposure.
Abbreviations: ANOVA, analysis of variance; SE, standard error.