Figures & data
Table 1 Summary of miRNAs That Promote or Antagonize HIV Latency
Figure 1 Regulation of HIV latency by cellular miRNAs. MiRNAs can regulate HIV latency via three distinct mechanisms. All rely on the binding of a cellular miRNA, in complex with the RNA-induced silencing complex (RISC), to the 3’ end of cellular or viral mRNAs. This either leads to translational repression or mRNA degradation. (Left) Some miRNAs reduce the levels of HIV activator proteins (ie transcriptional activators), leading to maintenance of the HIV latent reservoir. (Middle) Alternatively, other miRNAs decrease the expression of HIV repressor proteins, such as transcriptional silencers or restriction factors. This facilitates activation of the proviral LTR and latency reversal. (Right) MiRNAs may also promote latency post-transcriptionally via the repression of HIV spliced or unspliced transcripts, leading to reduced levels of HIV viral proteins. Specific cellular miRNAs and their cellular or viral mRNA target(s) are listed; miRNAs/target(s) that promote latency are highlighted in red, while those that have anti-latency properties are highlighted in green. The figure was created in Biorender.com.
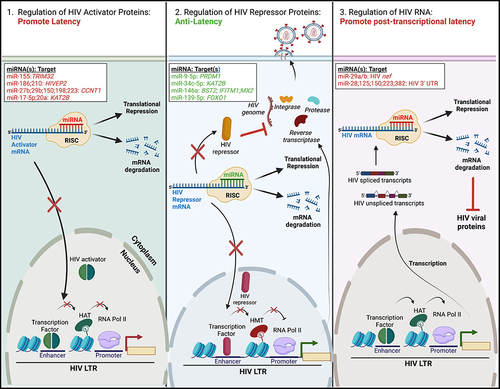
Table 2 Summary of lncRNAs That Promote or Antagonize HIV Latency
Figure 2 Regulation of HIV latency by cellular lncRNAs. Similar to miRNAs, lncRNAs may facilitate the regulation of HIV latency via three mechanisms, two of which exist at the epigenetic level. (Left) LncRNAs can bind and/or recruit HIV repressors to the HIV 5′ LTR. This leads to repressive epigenetic modifications, such as histone methylation. Alternatively, lncRNAs can inhibit the binding/recruitment of HIV activators to the HIV 5′ LTR. Both cases would lead to transcriptional repression and latency maintenance. (Middle) Alternatively, some lncRNAs may either inhibit (HIV repressors) or promote (HIV activators) the recruitment of protein complexes to the HIV-1 5′ LTR, resulting in gene activation, and reactivation of HIV from latency. (Right) Other lncRNAs may promote latency post-transcriptionally. In one scenario, lncRNAs can associate with nuclear proteins to form paraspeckles, or complexes that regulate mRNA export. Binding of unspliced HIV transcripts to paraspeckles can inhibit their nuclear export, decreasing viral protein synthesis. Alternatively, lncRNAs can link viral regulatory proteins (Tat) to the ubiquitin proteasome system, facilitating their premature degradation. Specific lncRNAs and their cellular or viral protein target(s) are listed; lncRNAs/targets that promote latency are highlighted in red, while those that have anti-latency properties are highlighted in green. The figure was created in Biorender.com.
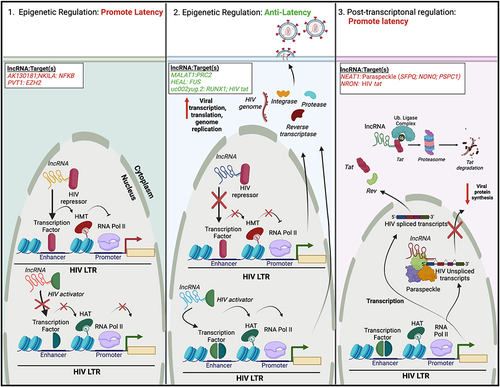
Table 3 Summary of RNAs Encoded in the HIV Genome That Promote or Antagonize HIV Latency
Figure 3 Regulation of HIV latency by viral RNAs. (Left) In some instances, viral transcription from the HIV LTR produces short non-coding HIV transcripts (termed TAR-gag). These viral transcripts can either be packaged into exosomes or recruit HIV repressor proteins to facilitate latency maintenance. (Middle) TAR-gag encodes viral pre-miRNAs that are processed by the cellular protein Dicer to form mature viral miRNAs. These viral miRNAs either facilitate protein degradation of HIV viral proteins or associate with HIV repressor proteins to mediate transcriptional silencing of the HIV LTR. Three additional virally encoded miRNAs have variable function in regulating latency. (Right) The HIV genome may transcribe antisense RNA. Like short HIV transcripts, these regulatory RNAs associate with HIV repressors to induce viral transcriptional silencing. Specific non-coding HIV RNAs and their target(s) are listed; HIV RNAs/targets that promote latency are highlighted in red, those that have anti-latency properties are highlighted in green, and those whose function in HIV latency is still unclear are highlighted in black. The figure was created in Biorender.com.
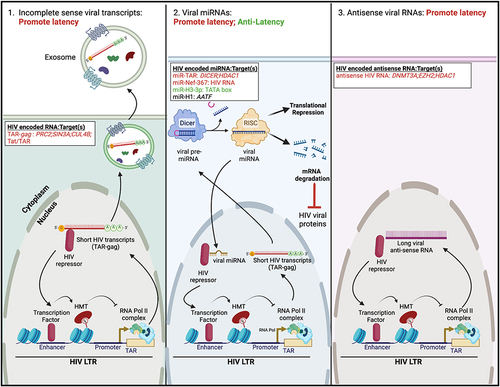
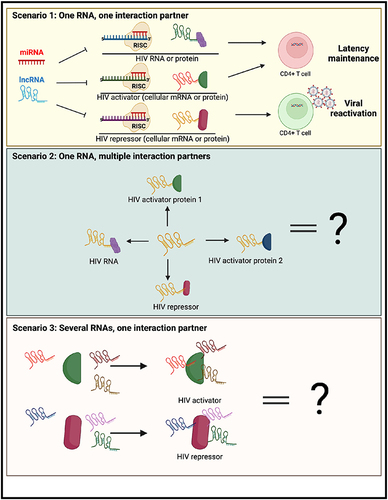
Figure 4 Summary of different scenarios for therapeutic targeting of regulatory RNAs for an HIV cure. (Top) The best-case scenario for an HIV cure would be the identification of a regulatory RNA, preferably a viral RNA, that has one interaction partner (HIV RNA, HIV activator or HIV repressor). Targeting of this RNA could lead to latency reversal and/or long-term proviral silencing. (Middle) Alternatively, a regulatory RNA may have diverse functions with different types of HIV regulators. (Bottom) Finally, regulators of HIV (activators/repressors) may interact with multiple regulatory RNAs, and thus disrupting their function may necessitate novel approaches. The figure was created in Biorender.com.