Figures & data
Table 1 Bacterial strains used in this study
Table 2 The primers used in this study
Figure 1 Location and sequence of the ctrA gene of Neisseria meningitidis applied for the MCDA primer design. The diagram displays the sequence and design of MCDA primers. The nucleotide sequence of the sense strand of the ctrA gene of N. meningitidis was shown. Right arrows and left arrows indicate sense and complementary sequences which were used in the present study, respectively.
Abbreviation: MCDA, multiple cross displacement amplification.
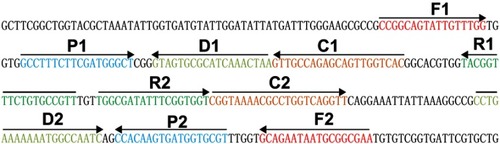
Figure 2 Confirmation and detection of Neisseria meningitidis-MCDA products. (A) By the MG method, amplification products of the N. meningitidis-MCDA assay were visually analyzed by observation of the color change. (B) A lateral flow biosensor was applied for visual detection of N. meningitidis-MCDA products. Tube 1/Biosensor 1: positive amplification of N. meningitidis strain 13007 Tube 2/Biosensor 2: negative control of Staphylococcus aureus (GZCDC isolate); Tube 3/Biosensor 3: negative control of Streptococcuspneumoniae (GZCDC isolate); Tube 4/Biosensor 4: blank control (DW).
Abbreviations: TL, test line; CL, control line; MCDA, multiple cross displacement amplification; MG, malachite green; GZCDC, Guizhou Provincial Center for Disease Control and Prevention; DW, double-distilled water.
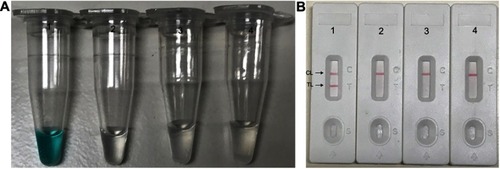
Figure 3 Reaction temperature optimization for N. meningitidis-MCDA primers. The standard MCDA reactions for detection of N. meningitidis were monitored by the determination of real-time turbidity, and the DNA concentrations were displayed with corresponding curves marked in the figures. The threshold value was 0.1 and a turbidity >0.1 was set as positive. A total of 8 kinetic graphs (A–H) were produced at different temperatures points (60–67°C, 1°C intervals) with target DNA at the level of 10 pg per reaction. The graphs from (B–H) show strong amplification.
Abbreviation: MCDA, multiple cross displacement amplification.
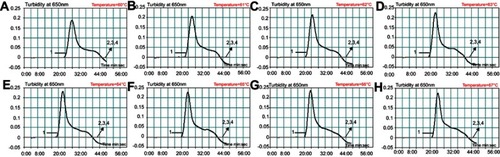
Figure 4 Sensitivity analysis of the MCDA-LFB assay using serial dilutions of genomic DNA extracted from Neisseriameningitidis strain 13007. A total of three monitoring techniques, including the lateral flow biosensor (A), real-time turbidity (B) and colorimetric indicator (MG; C) methods, were applied to analyze the amplification products. Serial dilutions of target templates were subjected to standard MCDA reactions. Biosensors (A)/Tubes (B)/Turbidity signals (C) 1–8 represent the DNA levels of 1 ng per reaction, 100 pg per reaction, 10 pg per reaction, 1 pg per reaction, 100 fg per reaction, 10 fg per reaction, 1 fg per reaction of target templates and the blank control (DW), respectively. The genomic DNA levels of 1 ng per reaction, 100 pg per reaction, 10 pg per reaction, 1 pg per reaction, 100 fg per reaction, 10 fg per reaction produced positive reactions.
Abbreviations: TL, test line; CL, control line; MCDA-LFB, multiple cross displacement amplification with lateral flow biosensor; MG, malachite green; DW, double-distilled water.
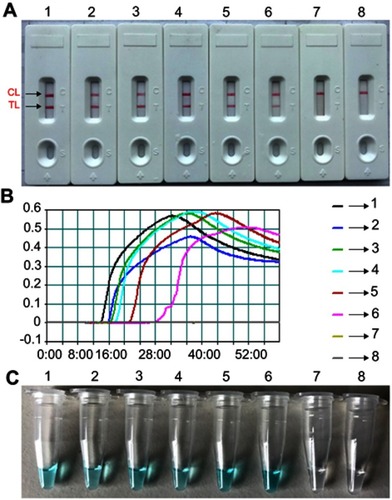
Figure 5 The optimal duration of time required for Neisseria meningitidis-MCDA-LFB method. Four distinct reaction times (A, 15 mins; B, 25 mins; C, 35 mins; D, 45 mins) were examined and compared at 64°C. Biosensors 1, 2, 3, 4, 5, 6, 7 and 8 represent DNA levels of 1 ng of templates, 100 pg of N. meningitidis templates, 10 pg of N. meningitidis templates, 1 pg of N. meningitidis templates, 100 fg N. meningitidis templates, 10 fg N. meningitidis template, 1 fg N. meningitidis template and blank control (DW), respectively. The best sensitivity was observed when the amplification lasted for 35 mins (C).
Abbreviations: TL, test line; CL, control line; MCDA-LFB, multiple cross displacement amplification with lateral flow biosensor; DW, double-distilled water.
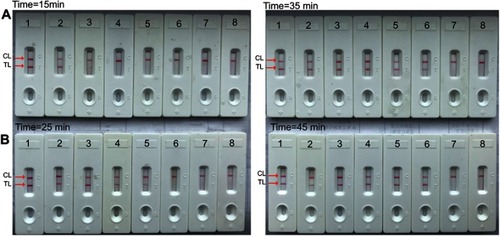
Figure 6 Specificity analysis of the Neisseriameningitidis-MCDA-LFB assay for different bacterial strains. MCDA reactions were carried out using different genomic DNA templates and were analyzed visually. Biosensors 1, 2, 3, 4, 5, 6, 7, 8, 9 used N. meningitidis serogroup A, N. meningitidis serogroup B, N. meningitidis serogroup C, N. meningitidis serogroup D, N. meningitidis serogroup W-135, N. meningitidis serogroup 29-E, N. meningitidis serogroup X, N. meningitidis serogroup Y, N. meningitidis serogroup Z; 10–11, Biosensors 10–11 used N. meningitidis isolate GZCDC001, N. meningitidis isolate GZCDC002, respectively; Biosensors 12–39 used Bordetella pertussis, Bordetella parapertussis, Hemophilus parainfluenza, Streptococcus pneumoniae, Klebsiella pneumoniae, Pseudomonas aeruginosa, Mycoplasma pneumoniae, Legionellae bacillus, Acinetobacter baumannii, Staphylococcus aureus, Staphylococcus saprophyticus, Salmonella, enteropathogenic E. coli, enterotoxigenic E. coli, invasive E.coli, enterohemorrhagic E. coli, enteroaggregative E. coli, Streptococcus suis, Vibrio cholerae, Vibrio parahemolyticus, Enterococcus faecalis, Enterococcus faecium, Bacillus cereus, Bacillus proteus, Enterobacter cloacae; Listeria monocytogenes, Shigella flexneri, Shigella boydii, respectively; Biosensor 40 used the blank control (DW).
Abbreviations: MCDA-LFB, multiple cross displacement amplification with lateral flow biosensor; DW, double-distilled water; GZCDC, Guizhou Provincial Center for Disease Control and Prevention.
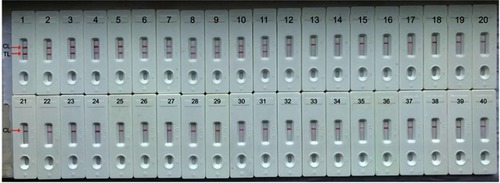
Table 3 Comparison of conventional PCR, culture-biotechnical and MCDA-LFB for the detection of N. meningitidis in clinical samples
