Figures & data
Table 1 Primers Used in This Study
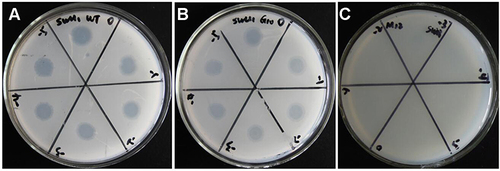
Figure 1 Sensitivity of wild-type Mycobacterium smegmatis mc2 155 and M12 to phage SWU1. (A) wild-type M. smegmatis mc2 155 strain can form obvious plaque for SWU1. (B) M12 mutant strain cannot form plaque.
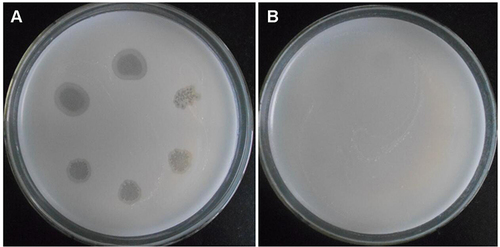
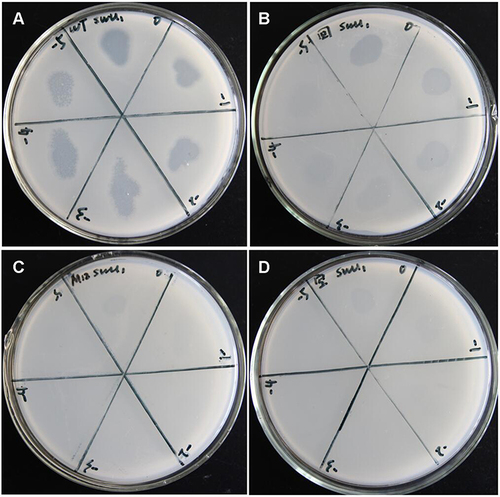
Figure 2 Detection of M12 resistance to SWU1. (A) As a blank control. (B) M12, and (C) M. smegmatis mc2 155 does not affect the replication of SWU1. Red arrows indicate plaques of phages.
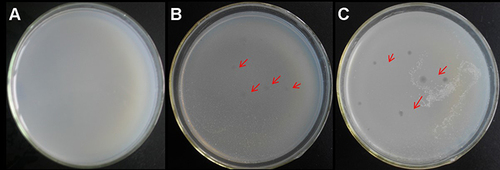
Figure 3 M12 mutant strain affects SWU1 adsorption to mycobacteria. (A and B) Wild-type M. smegmatis mc2 155 can adsorb SWU1 normally. (C and D) The adsorption capacity of M12 to SWU1 was significantly lower than that of M. smegmatis mc2 155. Red arrows indicate the location of phage presence.
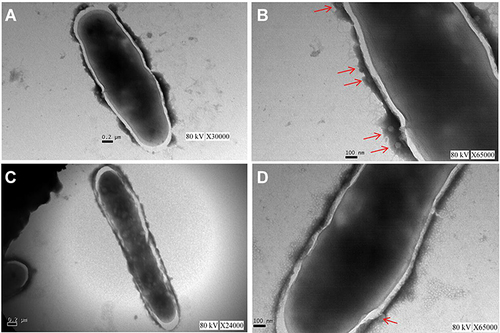
Figure 4 The cell wall of M12 mutant was changed. Single colony phenotype of (A) M12 mutant and (B) wild-type M. smegmatis mc2 155 strains. (C/D) The fatty acid analysis results of (C) M12 mutant strains and (D) wild-type M. smegmatis mc2 155. Red boxs indicate a significant change in the abundance of components of the content. (E) Changes of fatty acid content in cell wall of M12 mutant compared with Mycobacterium smegmatis MC2 155.
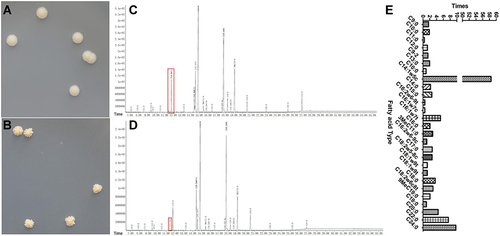
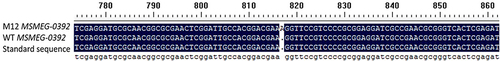
Figure 5 Transposon insertion site is located in the gene MSMEG_3705. (A) PCR amplification of MSMEG_3705 gene by specific primers. Lane 1: Marker DL8000. Lane 2/3: MSMEG_3705 amplification product using M12 as template. Lane 4/5: MSMEG_3705 amplification using M. smegmatis mc2 155 as template. Lane 6: Marker DL2000. (B) Schematic diagram of insertion of Tn5 transposon into M. smegmatis mc2 155. The asterisk indicates the transposon Tn5 insertion site.
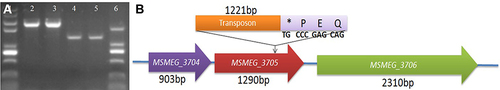
Figure 6 Sensitivity detection of different strains to SWU1. (A) M. smegmatis mc2 155 and (B) M. smegmatis mc2155 ∆MSMEG_3705 was sensitive to SWU1. (C) M12 mutant strain cannot form plaque.