Figures & data
Figure 1 Drug susceptibility heat map of 33 KPC-producing Klebsiella pneumoniae. The minimum inhibitory concentrations (MICs) of 33 KPC-producing Klebsiella pneumoniae strains were homogenized against 17 antibiotics, and the colors in the legend indicate the change in susceptibility of the strains to the antibiotics,kP-s18, KP-s29 and KP-s39 without carbapenemase resistance genes were indicated in gray.
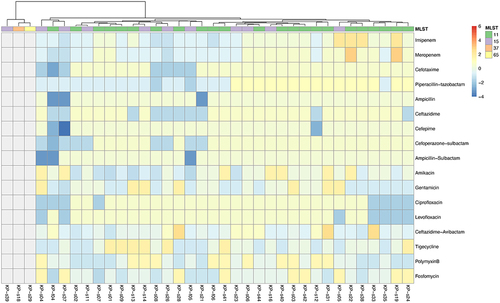
Figure 2 Comparative genomic circle map of 36 strains of Klebsiella pneumoniae. Taking KP-s26 as the reference genome, the remaining 35 Klebsiella pneumoniae were compared with KP-s26 for circle map.In the legend different colors represent different strains, black is GC content, green is GC offset of the leading chain, and purple is GC offset of lagging chain.Mark the missing genes on the circle map.
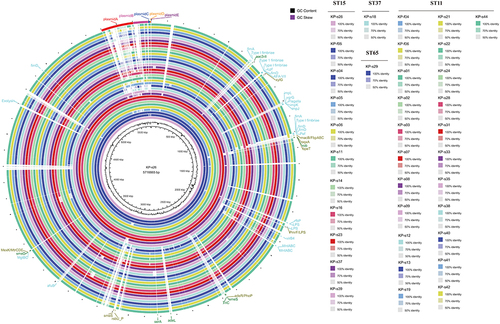
Table 1 Distribution of Drug Resistance Genes in 36 Strains of Klebsiella pneumoniae
Figure 3 Phylogenetic tree of 36 Klebsiella pneumoniae. Construction Phylogenetic tree of 36 Klebsiella pneumoniae strains.In the legend different colors were used to represent the ST typing of strains, the expression of blaKPC and blaCTX in the second and third layers of the figure, respectively.
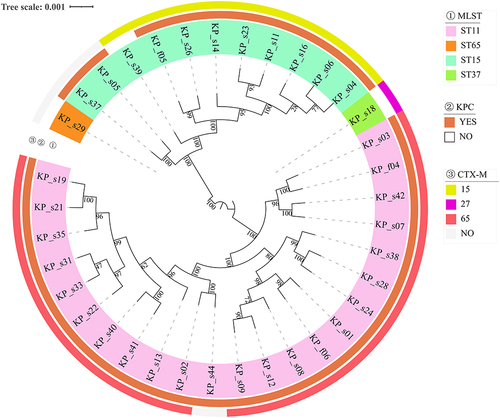
Figure 4 KPC gene sequence comparison of 33 KPC-producing Klebsiella pneumoniae. The figure shows the results of KPC gene sequence comparison among 33 KPC-producing Klebsiella pneumoniae strains. Different colors indicated the degree of sequence inconsistency, and black indicated that the sequence similarity was 100%. The KPC-24 gene carried by KP-s42 and KP-f04 is located at the edge of the scaffold, and the prediction is incomplete.

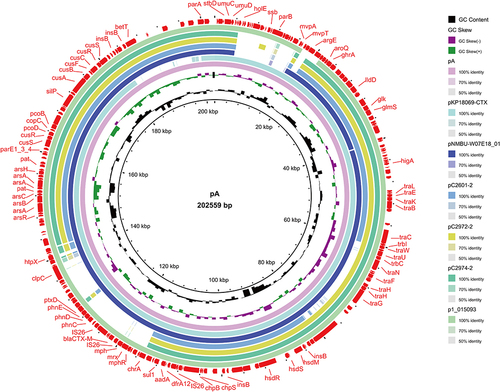
Figure 5 Plasmid map. Plasmid pA were compared with pKP18069-CTX, pNMBU-W07E18_01, pC2601-2, pC2972_2, pC2974_2, p1_015093, and plasmid pB uses MK191023.1 as the reference genome. In the outermost layer of the map, the drug resistance gene, movable element and type IV secretion system carried by the plasmid were marked. The missing portion indicates no expression or less than 50% genomic similarity in the strain genome compared to the reference genome.
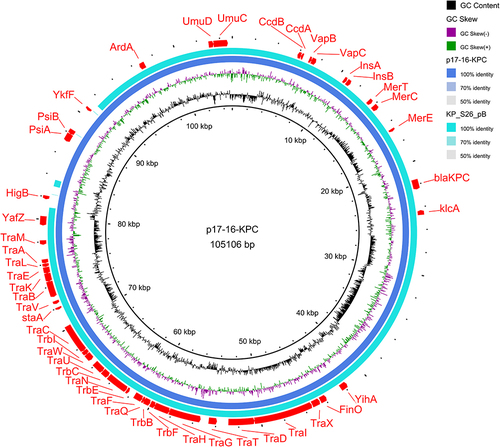
Table 2 2 Conjugate Plasmids in Klebsiella pneumoniae KP-S26