Figures & data
Table 1 Gender and Age Group of Blastocystis sp.-Infected and Uninfected Patients
Figure 1 Median joining network sequence analysis of SSU rRNA gene-PCR products demonstrated different haplotypes (H1-15) within Blastocystis sp. subtypes. Maximum parsimony (MP) analysis was used as the optional post-processing calculation using Network 4.6.1.3 software. Each haplotype represents a distinct subtype. The haplotypes are numbered from H1 to H15.
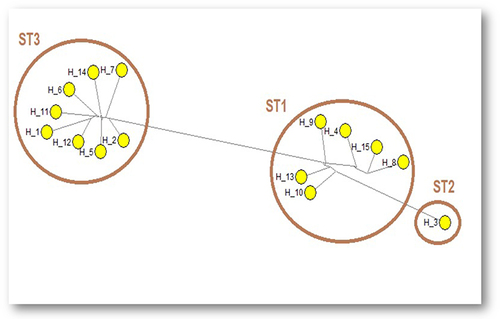
Table 2 Inter- and Intra-Subtype Genetic Variations Characterize Blastocystis sp. Subtypes/Haplotypes
Table 3 Genetic Variability of Blastocystis sp. Haplotypes
Table 4 GenBank References for the Blastocystis sp. Subtypes Used to Construct the Phylogenetic Tree
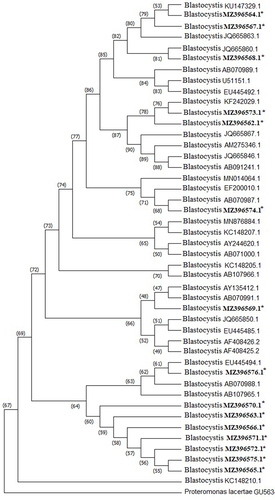
Figure 2 Phylogenetic relationship of Blastocystis sp. haplotypes identified in the current study to other Blastocystis sp. isolates, based on SSU rRNA gene sequences. The reference GenBank sequence accession numbers of all isolates are shown, including our haplotypes accession numbers in bold. Proteromonas lacertae served as the outgroup sequence. *Indicates the haplotypes identified in the current study.