Figures & data
Table 1 Antimicrobial Susceptibility Profiles of NDM-Producing Aeromonas caviae of Isolates
Figure 1 Construction of phylogenetic trees of A. caviae. The figure includes the sample source of the isolate, MLST, antibiotic resistance gene, and the comparison result of the virulence gene.
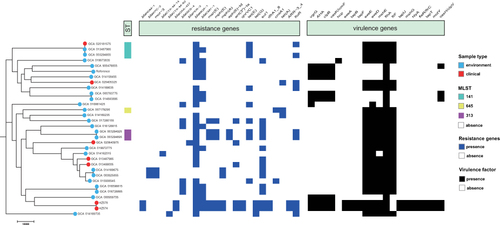
Figure 2 S1 Nuclease-pulsed field gel electrophoresis (S1-PFGE) and southern blot. The number and size of the plasmid of the two isolates were characterized by the S1 Nuclease-pulsed field gel electrophoresis (S1-PFGE).

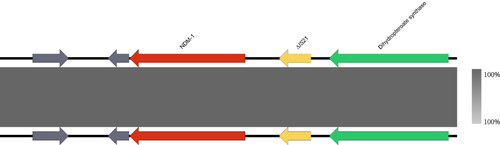
Figure 3 The genetic environment of the blaNDM-1 gene in A. caviae was isolated from clinical sources. The arrows represent the direction of transcription. The red open reading frame (ORF) indicates the blaNDM-1 gene, the yellow ORF indicates the mobile element, the green ORF indicates enzymes, and the grey ORF indicates other genes or genes of unknown function.