Figures & data
Figure 1 Chemical structure of berberine. Adapted from Ai X, Yu P, Peng L, et al. Berberine: a review of its pharmacokinetics properties and therapeutic potentials in diverse vascular diseases. Front Pharmacol. 2021;12:762654. Creative Commons.Citation24
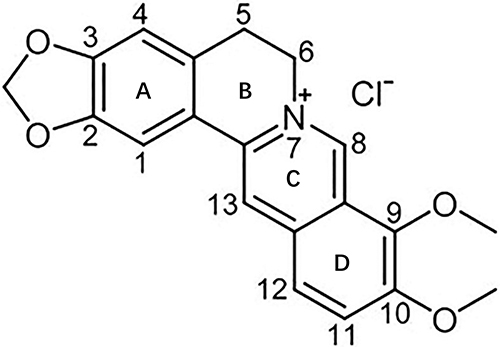
Table 1 The Effect of Sub-MIC Berberine on Antibiotics Against Common Multidrug-Resistant Bacteria
Figure 2 The synergistic mechanism of berberine (BER) in inhibiting antibiotic efflux. (A) BER inhibits the expression of efflux pump genes, preventing antibiotic efflux. In Pseudomonas aeruginosa, Acinetobacter xylosoxidans and Burkholderia cepacia, BER significantly reduces the expression of the MexXY multidrug efflux system, leading to a decrease in efflux of various antibiotics, including aminoglycosides (amikacin, arbekacin, gentamicin and tobramycin), β-lactams (cefepime and imipenem), macrolides (erythromycin), and lincosamides (lincomycin). (B) BER acts as a competitor in inhibiting antibiotic efflux. In Acinetobacter baumannii, BER enhances the expression of the AdeABC efflux pump gene adeB, and in multidrug-resistant Klebsiella pneumoniae, BER upregulates the expression of the acrA, acrB, tolC and acrR genes associated with the AcrAB-TolC efflux pump. BER exhibits a greater affinity for these efflux pumps and is preferentially pumped out, thereby reducing the efflux of other corresponding antibiotics. The figure was created with BioRender.com (https://app.biorender.com).
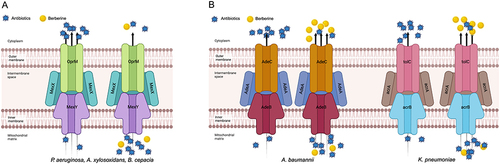
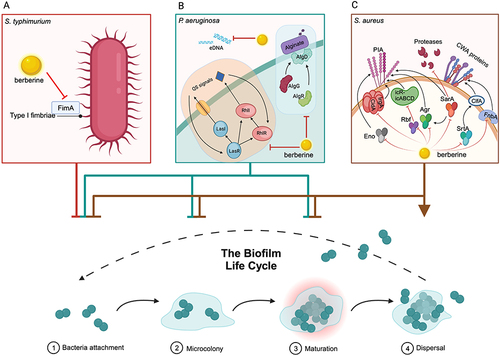
Figure 3 Berberine (BER) enhances antibiotic resistance by inhibiting the biofilm formation. (A) BER reduces the expression and quantity of type I fimbriae by affecting the expression of the fimA gene, thereby decreasing the activity and adhesion of Salmonella typhimurium. (B) BER affects the bacterial attachment, microcolony and biofilm maturation of Pseudomonas aeruginosa by reducing the levels of QS molecules and the expression of key genes involved in biofilm establishment and structural stability, such as lasI, lasR, rhlI, rhlR, eDNA and the alginate-related regulatory genes algG, algD and algR. (C) BER inhibits the attachment, microcolony formation and biofilm maturation, and promotes biofilm dispersal of Staphylococcus aureus by significantly downregulating the expression of genes associated with biofilm formation, such as srtA, agr, sarA, fnbA, rbf, lrgA, cidA and eno. These genes affect the production of polysaccharide intercellular adhesin (PIA), multiple extracellular proteases, and bacterial cell wall anchoring (CWA) proteins, etc. The figure was created with BioRender.com (https://app.biorender.com).