Figures & data
Table 1 Top 10 Up- and Down-Regulated DEmRNAs, DEmiRNAs and DElncRNAs
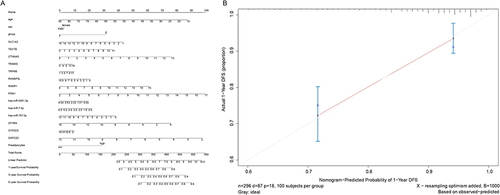
Figure 1 The hierarchical clustering results of top 100 up- and down-regulated DEmRNAs (A), all DEmiRNAs (B) and all DElncRNAs (C) between the early and late stage HCC. Row and column represented DEmRNAs/DEmiRNAs/DElncRNAs and samples, respectively. The color scale represented the expression levels. Orange indicates above the reference channel (high expression genes). Blue indicates below the reference channel (low expression genes).
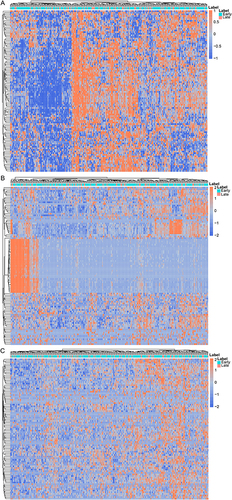
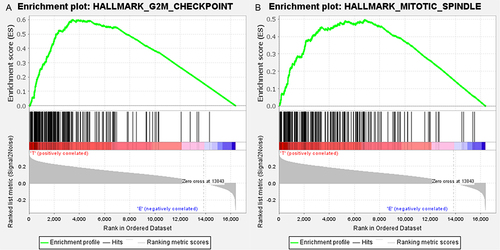
Figure 2 GSEA analysis illustrated up-regulated gene sets in the late stage HCC. (A) G2M_checkpoint; (B) mitotic_spindle.
Data Sharing Statement
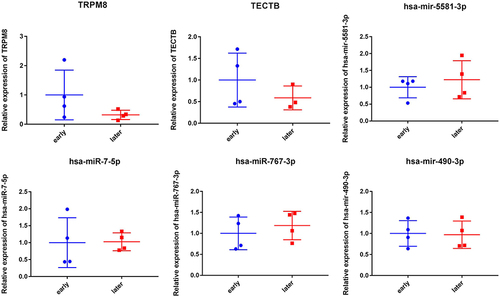
All data generated or analyzed during this study are included in this published article. We searched for HCC public gene expression data and complete clinical annotations from TCGA (https://tcga-data.nci.nih.gov/tcga/) database. The accession numbers is TCGA: HCC (Platform: Illumina RNAseq), respectively. The raw data of RT-qPCR validation has been deposited in the Harvard Dataverse database (https://doi.org/10.7910/DVN/I8L6QI).