Figures & data
Figure 1 Schematic of the AuNP-mediated Taqman-MGB nanoPCR for single-base mutation detection. When amplification of mutant-type and wild-type templates is mediated by the mutant-type probes, the AuNP particles inhibit the mismatch pairing of wild-type template with the probe and the corresponding amplification fluorescence signal is significantly suppressed. While the fluorescence signal generated from the amplification of mutant template is almost unaffected.
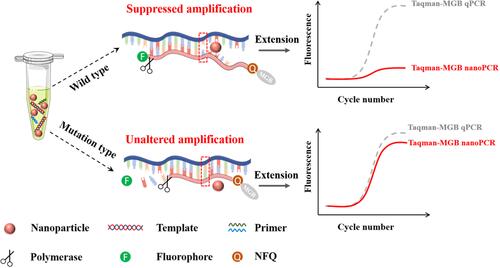
Figure 2 Effect of different AuNP concentrations on amplification signals. (A) The amplification curves of Taqman-MGB qPCR and nanoPCR with different AuNPs concentrations. The hollow square represents the amplification curve of wild-type templates, and the solid circle represents the amplification curve of mutant templates. (B) Comparison of the amplification single of WT and MT templates when different AuNPs concentrations are doped in Taqman-MGB nanoPCR. Mean fluorescence intensity and standard deviation are obtained at 45 cycle number with three replicate experiments. (C) Quantification of signal-to-noise of Taqman-MGB nanoPCR with different AuNP concentrations by calculating the amplification signal ratio of MT and WT templates.
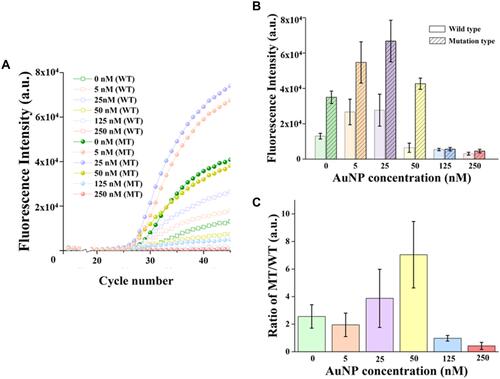
Figure 3 Mechanism of AuNPs for improving the specificity of Taqman-MGB nanoPCR. (A) The schematic of energy barrier hypothesis to explain the effect of AuNPs on the perfectly and mismatch complement pairing between templates and probes. (B) Hybridization of two single-stranded template sequences (wild-type and mutation-type) with MT probe at different AuNP particle concentrations. The fluorescent signal is derived from the double-stranded product with binding of SYBR Green I. (C) Average fluorescence intensities of WT templates and MT templates when the fluorescence signals reach steady state after hybridization.
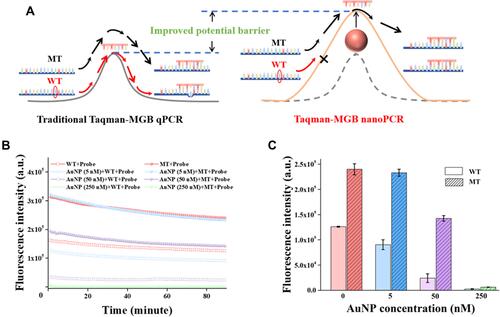
Figure 4 Specificity comparison of Taqman-MGB qPCR and nanoPCR at different target concentrations. The average fluorescence intensities of (A) Taqman-MGB qPCR and (B) nanoPCR at different target concentrations. The fluorescent intensity is extracted from the value at 45 amplification cycle number. (C) Discrimination comparison of Taqman-MGB qPCR and nanoPCR by Rdis with different target concentrations. (D) The specificity improvement of Taqman-MGB nanoPCR varied with different target concentrations.
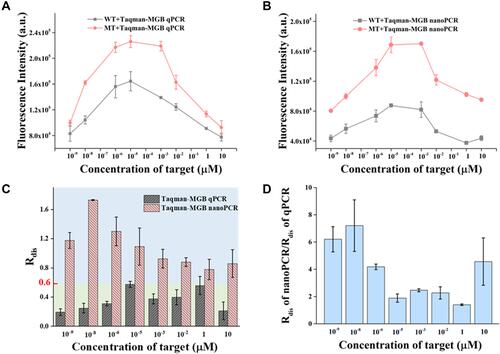
Table 1 LOD Comparison of Currently Reported qPCR Methods for T790M Mutation Abundance
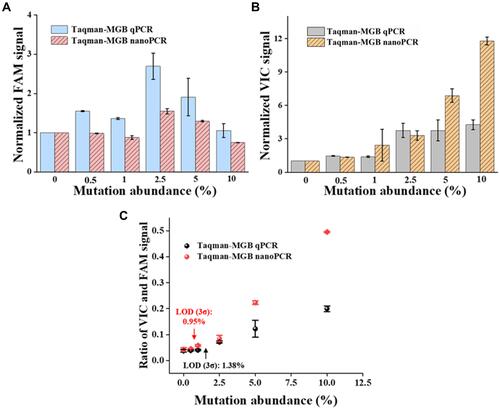
Figure 5 Detection ability comparison of Taqman-MGB qPCR and nanoPCR varied different mutation abundance. (A) The amplification signal of WT templates (ie, FAM fluorescence) varied with different mutation abundance. (B) The amplification signal of MT templates (ie, VIC fluorescence) varied with different mutation abundance. (C) Quantification of different mutation abundance by ratio of VIC and FAM signals in Taqman-MGB qPCR and nanoPCR. The fluorescence intensity is normalized to that of 0% mutation abundance group in figure A and B.