Figures & data
Figure 1 CircROBO1 is overexpressed in HCC. (A) Volcano plot showing differentially expressed circRNAs between tumor tissues and paired adjacent tissues from GSE156088 dataset. (B) Analysis of circROBO1 expression in 20 paired HCC tissues and adjacent tissues by qRT-PCR. (C) qRT-PCR analysis of circROBO1 expression in HCC cell lines and normal hepatic cell line LO2. (D) Schematic illustration showing the genomic loci of the ROBO1 gene and the circROBO1 is back-spliced from exon 4 to exon 6 of ROBO1. (E) qRT-PCR analysis for the resistance of circROBO1 and linear ROBO1 to RNase R in HCCLM9 and SMMC-7721 cells. The mock treatment is the negative control. (F) Actinomycin D assay to evaluate the stability of circROBO1 and ROBO1 mRNA in HCCLM9 and SMMC-7721 cells. (G) Nuclear-cytoplasmic fractionation assay was performed to examine the subcellular distribution of circROBO1 in HCCLM9 and SMMC-7721 cells. β-actin and U6 were used as cytoplasmic and nuclear positive controls, respectively. (H) Kaplan-Meier’s survival curves showing the correlations between circROBO1 expression and OS or RFS. Log rank test was used. Data were presented as means ±SD. **P<0.01; ***P<0.001.
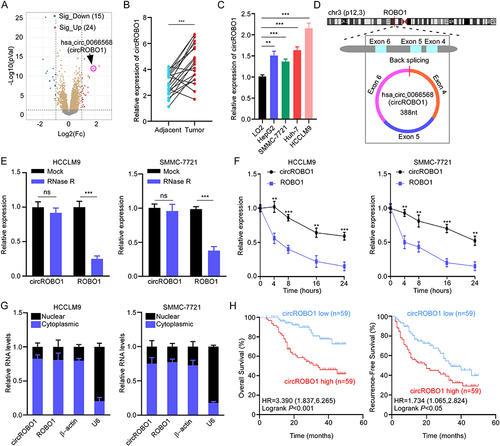
Figure 2 CircROBO1 overexpression promotes the proliferation of HCC cells. (A) The relative expression of circROBO1 in HCCLM9 and SMMC-7721 cells as indicated treatments was analyzed by qRT-PCR. (B–D) Cell proliferative ability was evaluated by CCK-8 (B), colony formation (C) and EdU (D) assays in HCCLM9 and SMMC-7721 cells as indicated treatments. Scale bar, 20 μm. (E) A xenograft tumor model was established via subcutaneously injecting circROBO1 silenced or overexpressed HCC cells. Tumor volume was measured at different time-points. Data were presented as means±SD. *P<0.05; **P<0.01; ***P<0.001.
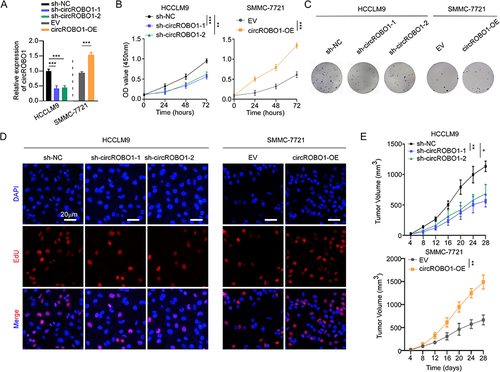
Figure 3 CircROBO1 functions as a sponge for miR-130a-5p. (A) Venn diagram showing potential target miRNAs of circROBO1 as predicted by Circbank and Starbase databases. (B) RNA pull-down with a biotin-labeled miR-130a-5p probe was conducted in HCC cell lines, the enrichment of circROBO1 was detected by qRT-PCR. (C) Schematic illustration of full length of wide type (WT) circROBO1 or a version where the miR-130a-5p-binding sites was mutated (MUT). (D) The luciferase activity of WT circROBO1 or MUT circROBO1 after transfection with miR-130a-5p mimic or inhibitor in HCCLM9 and SMMC-7721 cells, respectively. (E) RIP assay was performed to detect circROBO1 and miR-130a-5p binding to AGO2. (F) The expression of miR-130a-5p in HCC tissues and paired adjacent tissues from TCGA database. (G and H) The cell proliferative ability was detected in HCC cells as indicated treatments by colony formation (G) and EdU (H) assays. Data were presented as means ±SD. **P<0.01; ***P<0.001.
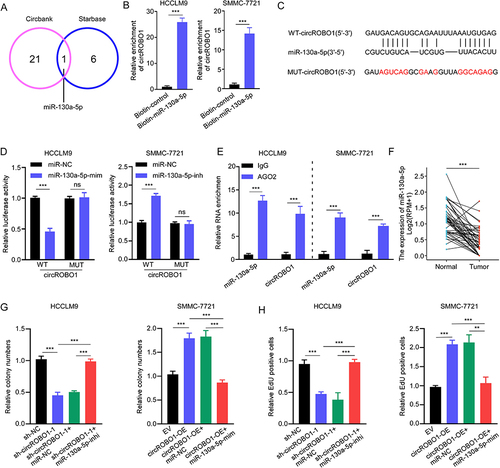
Figure 4 MiR-130a-5p inhibits HCC proliferation by negatively regulating CCNT2 expression. (A) Venn diagram showing the overlapping target genes of miR-130a-5p predicted by MicroT, miRDB and Starbase databases. (B) The expression of PTP4A2, CCNT2, and CFL2 in HCC tissues and paired adjacent tissues from TCGA database. (C) Pearson’s correlation analysis of CCNT2 and miR-130a-5p expression in 20 cases HCC tissues. (D) The relative expression of CCNT2 was detected in HCCLM9 and SMMC-7721 cells after transfection of miR-130a-5p mimic or miR-130a-5p inhibitor by qRT-PCR. (E) Schematic illustration of full length of CCNT2 3’-UTR wide type (WT) or a version where the miR-130a-5p binding sites were mutated (MUT). The luciferase activity of CCNT2 3′-UTR WT or CCNT2 3′-UTR MUT after transfection of miR-130a-5p mimics or inhibitor in HCCLM9 and SMMC-7721 cells, respectively. (F and G) The cell proliferative ability of HCCLM9 and SMMC-7721 cells as indicated treatments was detected by colony formation (F) and EdU (G) assays. Data were presented as means ±SD. *P<0.05; **P<0.01; ***P<0.001.
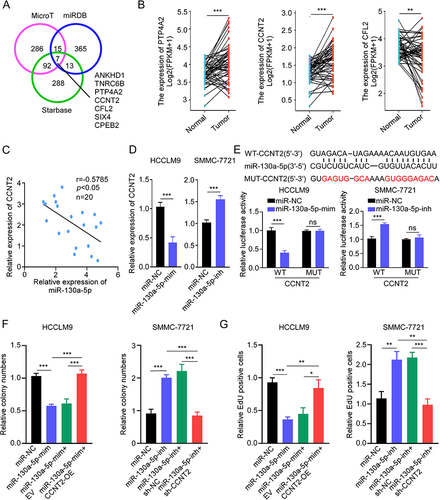
Figure 5 CircROBO1 promotes HCC proliferation by upregulating CCNT2 expression. (A) The relative expression of CCNT2 was detected in HCCLM9 and SMMC-7721 cells as indicated treatments. (B–D) The cell proliferative ability of HCCLM9 and SMMC-7721 cells as indicated treatments was detected by CCK8 (B), colony formation (C), and EdU (D) assays. Scale bar, 20 μm. Data were presented as means ±SD. **P<0.01; ***P<0.001.
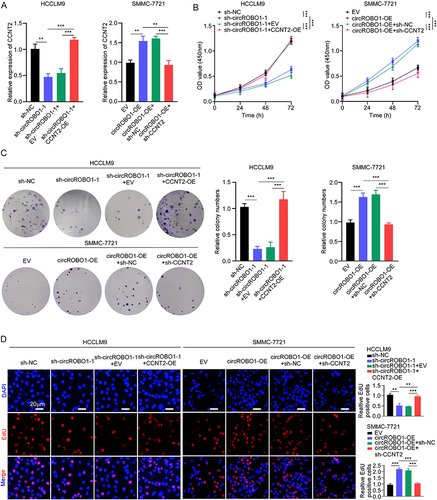
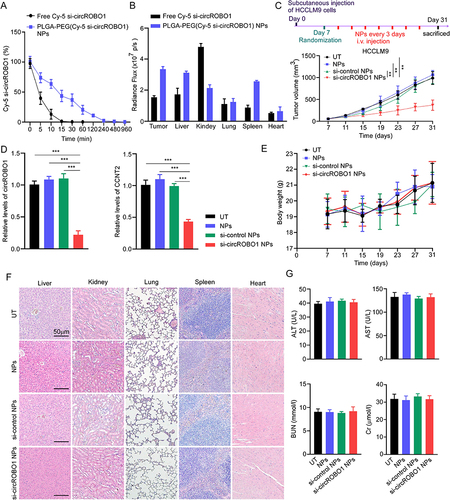
Figure 6 The biological characteristics of PLGA-PEG (si-circROBO1) NPs. (A) Schematic illustration of the PLGA-PEG (si-circROBO1) NPs. (B) Representative TEM image of PLGA-PEG (si-circROBO1) NPs. Scale bar, 200 nm. (C) Size distribution of the PLGA-PEG (si-circROBO1) NPs in the aqueous solution detected by Dynamic light scattering (DLS). (D) Zeta potential of PLGA-PEG (si-circROBO1) NPs in the aqueous solution . (E) In vitro stability of PLGA-PEG (si-circROBO1) NPs in 10% serum at 37°C was evaluated by measuring particle size changes with DLS at various time points up to 96 h. (F) The circROBO1 expression was determined by qRT-PCR in HCCLM9 cells which treated with PLGA-PEG (si-circROBO1) NPs at different siRNA doses. (G) The relative expression of circROBO1 was determined by qRT-PCR in HCCLM9 cells which treated with NPs, si-control NPs or si-circROBO1 NPs. (H) The proliferative ability of HCCLM9 cells which treated with NPs, si-control NPs or si-circROBO1 NPs was assessed by colony formation and EdU assays in vitro. Scale bar, 20 μm. Data were presented as means ±SD. **P<0.01; ***P<0.001.
Figure 7 Anti-tumor efficacy and toxicity evaluation of PLGA-PEG (si-circROBO1) NPs in vivo. (A) Blood circulation profile of free Cy-5 si-circROBO1 and PLGA-PEG (Cy-5 si-circROBO1) NPs. (B) Biodistribution of free Cy-5 si-circROBO1 and PLGA-PEG (Cy-5 si-circROBO1) NPs injections on major organs and tumors after 24 h of treatment. (C) Timeline of tumor implantation and treatment schedule in the HCC xenograft tumor-bearing nude mice. Tumor growth profile of the HCC xenograft tumor-bearing mice treated with NPs, si-control NPs and si-circROBO1 NPs. (D) The relative expression of circROBO1 and CCNT2 was evaluated by qRT-PCR in subcutaneous xenograft tumor tissues as indicated treatments. (E) Changes of mouse body weight during experimental period in subcutaneous xenograft tumor models. (F) Representative H&E staining of major organs including the liver, kidney, lung, spleen, and heart in subcutaneous xenograft tumor models, scale bar, 50 μm. (G) Blood ALT, AST, BUN, and Cr levels in subcutaneous xenograft tumor models. Data were presented as means ±SD. **P<0.01; ***P<0.001.