Figures & data
Figure 1 Schematic presentation showing the gene delivery by dual modified LNPs.
Notes: The nanoparticle is modified with angiopep for receptor-mediated uptake in LRP-1 expressing cells (left pathway) and a MMP-cleavable lipopeptide for activation in tumor tissue microenvironment (right pathway). Intra- or extracellular cleavage of the lipopeptide dePEGylates the LNP and reverses the surface charge from negative to positive leading to increased uptake and endosomal escape.
Abbreviations: LNPs, lipid nanoparticles; LRP-1, low-density lipoprotein receptor-related protein-1; PEG, poly(ethylene glycol).
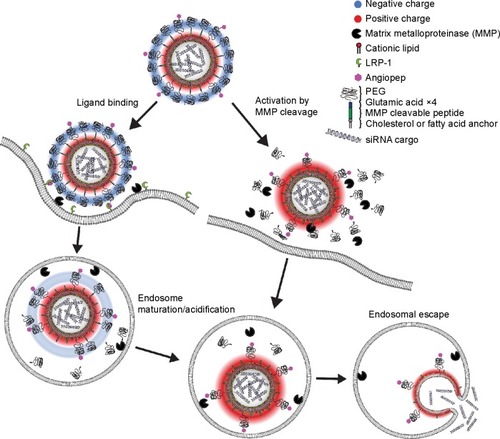
Table 1 Composition of LNPs expressed as mol%
Table 2 Characterization of LNPs by dynamic light scattering and PicoGreen assay
Figure 2 Uptake of angiopep-functionalized LNPs.
Notes: Representative histograms for bEnd.3 cells treated with A/PE-PEG-LNP (black), PE-PEG-LNP (gray), or buffer (dashed). Insert represents the MFI averaged over three samples. Error bars are SD.
Abbreviations: LNPs, lipid nanoparticles; PEG, poly(ethylene glycol); MFI, mean fluorescence intensity; SD, standard deviation.
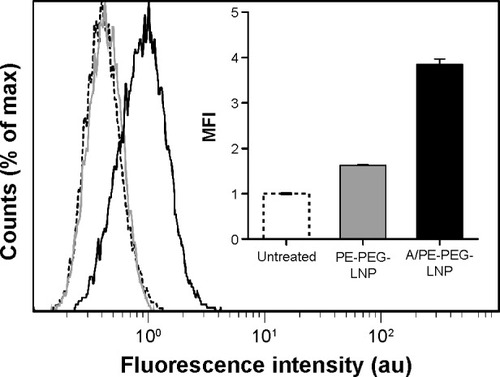
Figure 3 Proteinase cleaving of LNPs.
Notes: (A) The MALDI-TOF mass spectrum shows the size of the PEGylated lipids in PE-PEG-LNP, Chol-PCL-LNP, and DM-PCL-LNP when treated with buffer or thermolysin for cleavage of PCL. (B) ζ-potential of the LNPs when treated with buffer or thermolysin.
Abbreviations: LNPs, lipid nanoparticles; PEG, poly(ethylene glycol); PCL, PEGylated cleavable lipopeptide; Chol, cholesterol; DM, dimyristoyl; MALDI-TOF, matrix-assisted laser desorption/ionization time-of-flight.
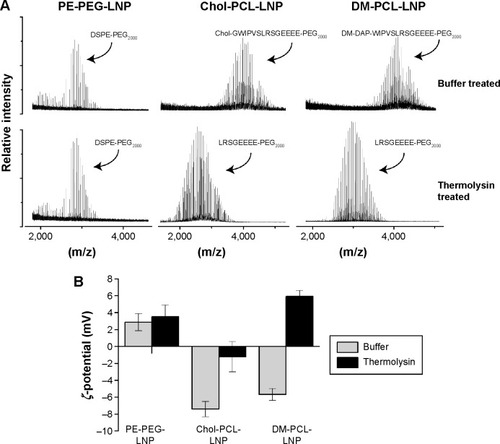
Figure 4 siRNA delivery with angiopep and Chol-PCL modified LNPs.
Notes: (A) Uptake of RhB-labeled LNPs in U87MG cells measured as fluorescence intensity of the cell lysate in arbitrary units (au). (B) Luciferase reporter activity relative to untreated cells after treatment with LNPs containing anti-luciferase siRNA. LNP dose corresponds to 120 nM siRNA. Error bars are SEM of two independent experiments performed in triplicates. *Significant difference from PE-PEG-LNP, **significant difference from Chol-PCL-LNP; determined using independent t-test P<0.05.
Abbreviations: PCL, PEGylated cleavable lipopeptide; LNPs, lipid nanoparticles; RhB, rhodamine B; PEG, poly(ethylene glycol); MMP, matrix metalloproteinase; SEM, standard error of mean; Chol, cholesterol.
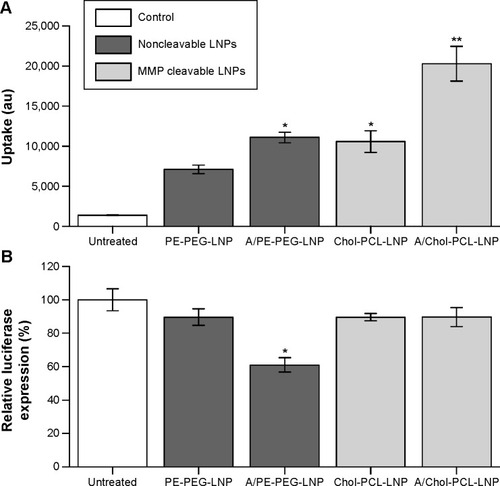
Figure 5 In vitro uptake of PE-PEG or DM-PCL containing LNPs in bEnd.3 and U87MG cells.
Notes: (A) Fraction of administered RhB-labeled lipid vehicle in the cell lysate. (B) Fraction of administered 33P labeled siRNA in the cell lysate. Error bars are SEM (n=4).
Abbreviations: PEG, poly(ethylene glycol); PCL, PEGylated cleavable lipopeptide; LNPs, lipid nanoparticles; RhB, rhodamine B; SEM, standard error of mean; DM, dimyristoyl.
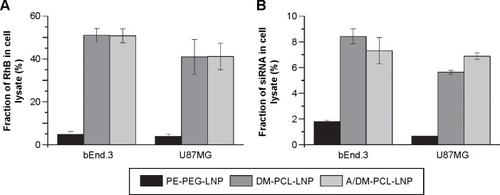
Figure 6 Knockdown of luciferase by LNPs.
Notes: (A) bEnd.3 and (B) U87MG cells were incubated for 48 hours with nanoparticles containing 60 (black), 120 (gray), or 240 (white) nM siRNA. The luciferase expression of the cells was normalized to their total protein contents and plotted as percentage of the expression level of nontreated cells. Nonsense siRNA (siGFP) and the commercial transfection agent RNAiMAX served as negative and positive control, respectively. Error bars are SEM (n=4).
Abbreviations: PEG, poly(ethylene glycol); PCL, PEGylated cleavable lipopeptide; LNPs, lipid nanoparticles; SEM, standard error of mean; DM, dimyristoyl.
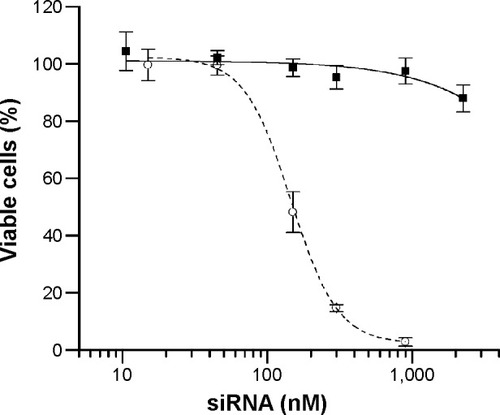
Figure 7 Cytotoxicity of DM-PCL-LNP.
Notes: MTS assay is used to analyze the proliferation of bEnd.3 cells treated with DM-PCL-LNP at siRNA concentrations ranging from 10 nM to 2.2 µM (squares). As control, cells treated with RNAiMAX (circles) were included. Error bars are SD.
Abbreviations: PCL, PEGylated cleavable lipopeptide; LNP, lipid nanoparticle; SD, standard deviation; DM, dimyristoyl.